Inter- and intragenus structural variations in caliciviruses and their functional implications
- PMID: 15163740
- PMCID: PMC416503
- DOI: 10.1128/JVI.78.12.6469-6479.2004
Inter- and intragenus structural variations in caliciviruses and their functional implications
Abstract
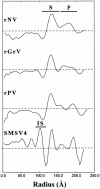
The family Caliciviridae is divided into four genera and consists of single-stranded RNA viruses with hosts ranging from humans to a wide variety of animals. Human caliciviruses are the major cause of outbreaks of acute nonbacterial gastroenteritis, whereas animal caliciviruses cause various host-dependent illnesses with a documented potential for zoonoses. To investigate inter- and intragenus structural variations and to provide a better understanding of the structural basis of host specificity and strain diversity, we performed structural studies of the recombinant capsid of Grimsby virus, the recombinant capsid of Parkville virus, and San Miguel sea lion virus serotype 4 (SMSV4), which are representative of the genera Norovirus (genogroup 2), Sapovirus, and Vesivirus, respectively. A comparative analysis of these structures was performed with that of the recombinant capsid of Norwalk virus, a prototype member of Norovirus genogroup 1. Although these capsids share a common architectural framework of 90 dimers of the capsid protein arranged on a T=3 icosahedral lattice with a modular domain organization of the subunit consisting of a shell (S) domain and a protrusion (P) domain, they exhibit distinct differences. The distally located P2 subdomain of P shows the most prominent differences both in shape and in size, in accordance with the observed sequence variability. Another major difference is in the relative orientation between the S and P domains, particularly between those of noroviruses and other caliciviruses. Despite being a human pathogen, the Parkville virus capsid shows more structural similarity to SMSV4, an animal calicivirus, suggesting a closer relationship between sapoviruses and animal caliciviruses. These comparative structural studies of caliciviruses provide a functional rationale for the unique modular domain organization of the capsid protein with an embedded flexibility reminiscent of an antibody structure. The highly conserved S domain functions to provide an icosahedral scaffold; the hypervariable P2 subdomain may function as a replaceable module to confer host specificity and strain diversity; and the P1 subdomain, located between S and P2, provides additional fine-tuning to position the P2 subdomain.
Figures






References
-
- Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. - PubMed
-
- Brünger, A. T. 1993. XPLOR manual,version 3.1. Yale University, New Haven, Conn.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

