Identification of three novel RB1 mutations in Brazilian patients with retinoblastoma by "exon by exon" PCR mediated SSCP analysis
- PMID: 15166261
- PMCID: PMC1770321
- DOI: 10.1136/jcp.2003.014423
Identification of three novel RB1 mutations in Brazilian patients with retinoblastoma by "exon by exon" PCR mediated SSCP analysis
Abstract
Aims: To carry out a retrospective study, screening for mutations of the entire coding region of RB1 and adjacent intronic regions in patients with retinoblastoma.
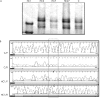
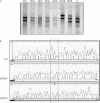
Methods: Mutation screening in DNA extracts of formalin fixed, paraffin wax embedded tissues of 28 patients using combined "exon by exon" polymerase chain reaction mediated single strand conformational polymorphism analysis, followed by DNA sequencing.
Results: Eleven mutations were found in 10 patients. Ten mutations consisted of single base substitutions; 10 were localised in exonic regions (eight nonsense, one missense, and one frameshift) and another one in the intron-exon splicing region. Three novel mutations were identified: a 2 bp insertion in exon 2 (g.5506-5507insAG, R73fsX77), a G to A transition affecting the last invariant nucleotide of intron 13 (g.76429G>A), and a T to C transition in exon 20 (g.156795T>C, L688P). In addition, eight C to T transitions, resulting in stop codons, were found in five different CGA codons (g.64348C>T, g.76430C>T, g.78238C>T, g.78250C>T, and g.150037C>T). Although specific mutation hotspots have not been identified in the literature, eight of the 11 mutations occurred in CGA codons and seven fell within the E1A binding domains (codons 393-572 and 646-772), whereas five were of both types-in CGA codons within E1A binding domains.
Conclusions: CGA codons and E1A binding domains are apparently more frequent mutational targets and should be initially screened in patients with retinoblastoma. Paraffin wax embedded samples proved to be valuable sources of DNA for retrospective studies, providing useful information for genetic counselling.
Figures
References
-
- Schultz KR, Ranade S, Neglia JP, et al. An increased relative frequency of retinoblastoma at a rural regional referral hospital in Miraj, Maharashtra, India. Cancer 1993;72:282–6. - PubMed
-
- Vogel F. Genetics of retinoblastoma. Hum Genet 1979;52:1–54. - PubMed
-
- Gallie BL, Moore A. Retinoblastoma. In: Taylor D, ed. Paediatric ophthalmology. Oxford: Blackwell Scientific, 1997:519–53.
-
- Yandell DW, Campbell TA, Dayton SH, et al. Oncogenic point mutations in the human retinoblastoma gene: their application to genetic counseling. N Engl J Med 1989;321:1689–95. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
