Integrative analysis of cell cycle control in budding yeast
- PMID: 15169868
- PMCID: PMC491841
- DOI: 10.1091/mbc.e03-11-0794
Integrative analysis of cell cycle control in budding yeast
Abstract
The adaptive responses of a living cell to internal and external signals are controlled by networks of proteins whose interactions are so complex that the functional integration of the network cannot be comprehended by intuitive reasoning alone. Mathematical modeling, based on biochemical rate equations, provides a rigorous and reliable tool for unraveling the complexities of molecular regulatory networks. The budding yeast cell cycle is a challenging test case for this approach, because the control system is known in exquisite detail and its function is constrained by the phenotypic properties of >100 genetically engineered strains. We show that a mathematical model built on a consensus picture of this control system is largely successful in explaining the phenotypes of mutants described so far. A few inconsistencies between the model and experiments indicate aspects of the mechanism that require revision. In addition, the model allows one to frame and critique hypotheses about how the division cycle is regulated in wild-type and mutant cells, to predict the phenotypes of new mutant combinations, and to estimate the effective values of biochemical rate constants that are difficult to measure directly in vivo.
Figures


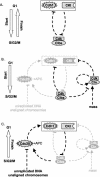


References
-
- Allen, N.A., et al. (2003). Modeling regulatory networks at Virginia Tech. Omics 7, 285-299. - PubMed
-
- Amon, A., Tyers, M., Futcher, B., and Nasmyth, K. (1993). Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74, 993-1007. - PubMed
-
- Bardin, A.J., and Amon, A. (2001). MEN and SIN: what's the difference? Nat. Rev. Mol. Cell. Biol. 2, 815-826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

