Genome-wide analysis of the biology of stress responses through heat shock transcription factor
- PMID: 15169889
- PMCID: PMC419887
- DOI: 10.1128/MCB.24.12.5249-5256.2004
Genome-wide analysis of the biology of stress responses through heat shock transcription factor
Abstract
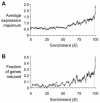
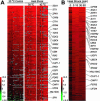
Heat shock transcription factor (HSF) and the promoter heat shock element (HSE) are among the most highly conserved transcriptional regulatory elements in nature. HSF mediates the transcriptional response of eukaryotic cells to heat, infection and inflammation, pharmacological agents, and other stresses. While HSF is essential for cell viability in Saccharomyces cerevisiae, oogenesis and early development in Drosophila melanogaster, extended life span in Caenorhabditis elegans, and extraembryonic development and stress resistance in mammals, little is known about its full range of biological target genes. We used whole-genome analyses to identify virtually all of the direct transcriptional targets of yeast HSF, representing nearly 3% of the genomic loci. The majority of the identified loci are heat-inducibly bound by yeast HSF, and the target genes encode proteins that have a broad range of biological functions including protein folding and degradation, energy generation, protein trafficking, maintenance of cell integrity, small molecule transport, cell signaling, and transcription. This genome-wide identification of HSF target genes provides novel insights into the role of HSF in growth, development, disease, and aging and in the complex metabolic reprogramming that occurs in all cells in response to stress.
Figures




References
-
- Amoros, M., and F. Estruch. 2001. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 39:1523-1532. - PubMed
-
- Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33:274-283. - PubMed
-
- Cahill, C. M., W. R. Waterman, Y. Xie, P. E. Auron, and S. K. Calderwood. 1996. Transcriptional repression of the prointerleukin 1β gene by heat shock factor 1. J. Biol. Chem. 271:24874-24879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
