RB reversibly inhibits DNA replication via two temporally distinct mechanisms
- PMID: 15169903
- PMCID: PMC419877
- DOI: 10.1128/MCB.24.12.5404-5420.2004
RB reversibly inhibits DNA replication via two temporally distinct mechanisms
Abstract
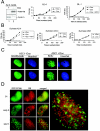
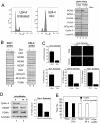
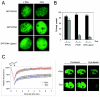
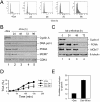
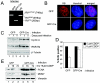
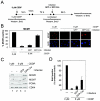
The retinoblastoma (RB) tumor suppressor is a critical negative regulator of cellular proliferation. Repression of E2F-dependent transcription has been implicated as the mechanism through which RB inhibits cell cycle progression. However, recent data have suggested that the direct interaction of RB with replication factors or sites of DNA synthesis may contribute to its ability to inhibit S phase. Here we show that RB does not exert a cis-acting effect on DNA replication. Furthermore, the localization of RB was distinct from replication foci in proliferating cells. While RB activation strongly attenuated the RNA levels of multiple replication factors, their protein expression was not diminished coincident with cell cycle arrest. During the first 24 h of RB activation, components of the prereplication complex, initiation factors, and the clamp loader complex (replication factor C) remained tethered to chromatin. In contrast, the association of PCNA and downstream components of the processive replication machinery was specifically disrupted. This signaling from RB occurred in a manner dependent on E2F-mediated transcriptional repression. Following long-term activation of RB, we observed the attenuation of multiple replication factors, the complete cessation of DNA synthesis, and impaired replicative capacity in vitro. Therefore, functional distinctions exist between the "chronic" RB-mediated arrest state and the "acute" arrest state. Strikingly, attenuation of RB activity reversed both acute and chronic replication blocks. Thus, continued RB action is required for the maintenance of two kinetically and functionally distinct modes of replication inhibition.
Figures




References
-
- Angus, S. P., A. F. Fribourg, M. P. Markey, S. L. Williams, H. F. Horn, J. DeGregori, T. F. Kowalik, K. Fukasawa, and E. S. Knudsen. 2002. Active RB elicits late G1/S inhibition. Exp. Cell Res. 276:201-213. - PubMed
-
- Avni, D., H. Yang, F. Martelli, F. Hofmann, W. M. ElShamy, S. Ganesan, R. Scully, and D. M. Livingston. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12:735-746. - PubMed
-
- Bartek, J., J. Bartkova, and J. Lukas. 1997. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 237:1-6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
