The J-protein family: modulating protein assembly, disassembly and translocation
- PMID: 15170475
- PMCID: PMC1299080
- DOI: 10.1038/sj.embor.7400172
The J-protein family: modulating protein assembly, disassembly and translocation
Abstract
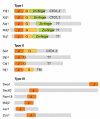
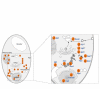
DnaJ is a molecular chaperone and the prototypical member of the J-protein family. J proteins are defined by the presence of a J domain that can regulate the activity of 70-kDa heat-shock proteins. Sequence analysis on the genome of Saccharomyces cerevisiae has revealed 22 proteins that establish four distinguishing structural features of the J domain: predicted helicity in segments I-IV, precisely placed interhelical contact residues, a lysine-rich surface on helix II and placement of the diagnostic sequence HPD between the predicted helices II and III. We suggest that this definition of the J-protein family could be used for other genome-wide studies. In addition, three J-like proteins were identified in yeast that contain regions closely resembling a J domain, but in which the HPD motif is non-conservatively replaced. We suggest that J-like proteins might function to regulate the activity of bona fide J proteins during protein translocation, assembly and disassembly.
Figures
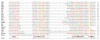
References
-
- Brodsky JL, Lawrence JG, Caplan AJ (1998) Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry 37: 18045–18055 - PubMed
-
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 - PubMed
-
- Craig EA, Eisenman HC, Hundley HA (2003) Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol 6: 157–162 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

