A core catalytic domain of the TyrA protein family: arogenate dehydrogenase from Synechocystis
- PMID: 15171683
- PMCID: PMC1133941
- DOI: 10.1042/BJ20031809
A core catalytic domain of the TyrA protein family: arogenate dehydrogenase from Synechocystis
Abstract
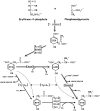
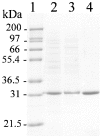
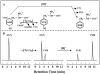
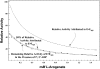
The TyrA protein family includes prephenate dehydrogenases, cyclohexadienyl dehydrogenases and TyrA(a)s (arogenate dehydrogenases). tyrA(a) from Synechocystis sp. PCC 6803, encoding a 30 kDa TyrA(a) protein, was cloned into an overexpression vector in Escherichia coli. TyrA(a) was then purified to apparent homogeneity and characterized. This protein is a model structure for a catalytic core domain in the TyrA superfamily, uncomplicated by allosteric or fused domains. Competitive inhibitors acting at the catalytic core of TyrA proteins are analogues of any accepted cyclohexadienyl substrate. The homodimeric enzyme was specific for L-arogenate (K(m)=331 microM) and NADP+ (K(m)=38 microM), being unable to substitute prephenate or NAD+ respectively. L-Tyrosine was a potent inhibitor of the enzyme (K(i)=70 microM). NADPH had no detectable ability to inhibit the reaction. Although the mechanism is probably steady-state random order, properties of 2',5'-ADP as an inhibitor suggest a high preference for L-arogenate binding first. Comparative enzymology established that both of the arogenate-pathway enzymes, prephenate aminotransferase and TyrA(a), were present in many diverse cyanobacteria and in a variety of eukaryotic red and green algae.
Figures

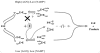

References
-
- Todd A. E., Orengo C. A., Thornton J. M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. - PubMed
-
- Teichmann S. A., Rison S. C. G., Thornton J. M., Riley M., Gough J., Clothia C. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J. Mol. Biol. 2001;311:693–708. - PubMed
-
- Stenmark S. L., Pierson D. L., Glover G. I., Jensen R. A. Blue–green bacteria synthesize L-tyrosine by the pretyrosine pathway. Nature (London) 1974;247:290–292. - PubMed
-
- Patel N., Pierson D. L., Jensen R. A. Dual enzymatic routes to L-tyrosine and L-phenylalanine via pretyrosine in Pseudomonas aeruginosa. J. Biol. Chem. 1977;252:5839–5846. - PubMed
-
- Zamir L. O., Jensen R. A., Arison B., Douglas A., Albers-Schonberg G., Bowen J. R. Structure of arogenate (pretyrosine), an amino acid intermediate of aromatic biosynthesis. J. Am. Chem. Soc. 1980;102:4499–4504.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

