PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings
- PMID: 15172342
- PMCID: PMC7106425
- DOI: 10.1016/S1473-3099(04)01044-8
PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings
Abstract
Molecular diagnostics are revolutionising the clinical practice of infectious disease. Their effects will be significant in acute-care settings where timely and accurate diagnostic tools are critical for patient treatment decisions and outcomes. PCR is the most well-developed molecular technique up to now, and has a wide range of already fulfilled, and potential, clinical applications, including specific or broad-spectrum pathogen detection, evaluation of emerging novel infections, surveillance, early detection of biothreat agents, and antimicrobial resistance profiling. PCR-based methods may also be cost effective relative to traditional testing procedures. Further advancement of technology is needed to improve automation, optimise detection sensitivity and specificity, and expand the capacity to detect multiple targets simultaneously (multiplexing). This review provides an up-to-date look at the general principles, diagnostic value, and limitations of the most current PCR-based platforms as they evolve from bench to bedside.
Figures


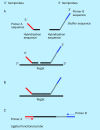
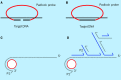
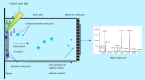
References
-
- Centers for Disease Control and Prevention Increase in national hospital discharge survey rates for septicemia—United States. MMWR Morb Mortal Wkly Rep. 1990;39:31–34. - PubMed
-
- Sands KE, Bates DW, Lanken PN. Epidemiology of sepsis syndrome in 8 academic medical centers. JAMA. 1997;278:234–240. - PubMed
-
- Pavlin JA, Gilchrist MJR, Osweiler GD. Diagnostic analyses of biological agent-caused syndromes: laboratory and technical assistance. Emerg Med Clin North Am. 2002;20:331–350. - PubMed
-
- Gerberding JL. Faster…but fast enough? Responding to the epidemic of severe acute respiratory syndrome. N Engl J Med. 2003;348:2030–2031. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

