Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli
- PMID: 15173110
- PMCID: PMC419781
- DOI: 10.1101/gr.2231904
Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli
Abstract
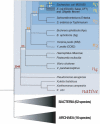
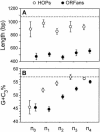
Differences in gene repertoire among bacterial genomes are usually ascribed to gene loss or to lateral gene transfer from unrelated cellular organisms. However, most bacteria contain large numbers of ORFans, that is, annotated genes that are restricted to a particular genome and that possess no known homologs. The uniqueness of ORFans within a genome has precluded the use of a comparative approach to examine their function and evolution. However, by identifying sequences unique to monophyletic groups at increasing phylogenetic depths, we can make direct comparisons of the characteristics of ORFans of different ages in the Escherichia coli genome, and establish their functional status and evolutionary rates. Relative to the genes ancestral to gamma-Proteobacteria and to those genes distributed sporadically in other prokaryotic species, ORFans in the E. coli lineage are short, A+T rich, and evolve quickly. Moreover, most encode functional proteins. Based on these features, ORFans are not attributable to errors in gene annotation, limitations of current databases, or to failure of methods for detecting homology. Rather, ORFans in the genomes of free-living microorganisms apparently derive from bacteriophage and occasionally become established by assuming roles in key cellular functions.
Copyright 2004 Cold Spring Harbor Laboratory Press
Figures


References
-
- Amiri, H., Davids, W., and Andersson, S.G. 2003. Birth and death of orphan genes in rickettsia. Mol. Biol. Evol. 20: 1575–1587. - PubMed
-
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474. - PubMed
-
- Bubunenko, M.G. and Subramanian, A.R. 1994. Recognition of novel and divergent higher plant chloroplast ribosomal proteins by Escherichia coli ribosome during in vivo assembly. J. Biol. Chem. 269: 18223–18231. - PubMed
-
- Charlebois, R.L., Clarke, G.D., Beiko, R.G., and St Jean, A. 2003. Characterization of species-specific genes using a flexible, web-based querying system. FEMS Microbiol. Lett. 225: 213–220. - PubMed
WEB SITE REFERENCES
-
- http://globin.cse.psu.edu/enterix; Percent Identity Plots on the EnteriX server.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
