Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales
- PMID: 15173603
- PMCID: PMC420407
- DOI: 10.1073/pnas.0401561101
Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales
Abstract
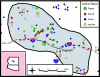
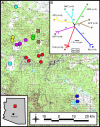
Plague, the disease caused by the bacterium Yersinia pestis, has greatly impacted human civilization. Y. pestis is a successful global pathogen, with active foci on all continents except Australia and Antarctica. Because the Y. pestis genome is highly monomorphic, previous attempts to characterize the population genetic structure within a single focus have been largely unsuccessful. Here we report that highly mutable marker loci allow determination of Y. pestis population genetic structure and tracking of transmission patterns at two spatial scales within a single focus. In addition, we found that in vitro mutation rates for these loci are similar to those observed in vivo, which allowed us to develop a mutation-rate-based model to examine transmission mechanisms. Our model suggests there are two primary components of plague ecology: a rapid expansion phase for population growth and dispersal followed by a slower persistence phase. This pattern seems consistent across local, regional, and even global scales.
Figures
References
-
- Poland, J. D. & Barnes, A. M. (1979) in CRC Handbook Series in Zoonoses, ed. Steele, J. (CRC, Boca Raton, FL), pp. 515-559.
-
- Link, V. B. (1955) Public Health Mon. 26, 1-120. - PubMed
-
- Biggins, D. E. & Kosoy, M. Y. (2001) J. Mammal. 82, 906-916.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

