Genome SEGE: a database for 'intronless' genes in eukaryotic genomes
- PMID: 15175116
- PMCID: PMC434494
- DOI: 10.1186/1471-2105-5-67
Genome SEGE: a database for 'intronless' genes in eukaryotic genomes
Abstract
Background: A number of completely sequenced eukaryotic genome data are available in the public domain. Eukaryotic genes are either 'intron containing' or 'intronless'. Eukaryotic 'intronless' genes are interesting datasets for comparative genomics and evolutionary studies. The SEGE database containing a collection of eukaryotic single exon genes is available. However, SEGE is derived using GenBank. The redundant, incomplete and heterogeneous qualities of GenBank data are a bottleneck for biological investigation in comparative genomics and evolutionary studies. Such studies often require representative gene sets from each genome and this is possible only by deriving specific datasets from completely sequenced genome data. Thus Genome SEGE, a database for 'intronless' genes in completely sequenced eukaryotic genomes, has been constructed.
Availability: http://sege.ntu.edu.sg/wester/intronless
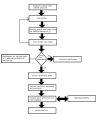
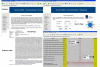
Description: Eukaryotic 'intronless' genes are extracted from nine completely sequenced genomes (four of which are unicellular and five of which are multi-cellular). The complete dataset is available for download. Data subsets are also available for 'intronless' pseudo-genes. The database provides information on the distribution of 'intronless' genes in different genomes together with their length distributions in each genome. Additionally, the search tool provides pre-computed PROSITE motifs for each sequence in the database with appropriate hyperlinks to InterPro. A search facility is also available through the web server.
Conclusions: The unique features that distinguish Genome SEGE from SEGE is the service providing representative 'intronless' datasets for completely sequenced genomes. 'Intronless' gene sets available in this database will be of use for subsequent bio-computational analysis in comparative genomics and evolutionary studies. Such analysis may help to revisit the original genome data for re-examination and re-annotation.
Figures
References
-
- Gilbert W. Why genes in pieces? Nature. 1978;271:501. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

