The loss of circadian PAR bZip transcription factors results in epilepsy
- PMID: 15175240
- PMCID: PMC423191
- DOI: 10.1101/gad.301404
The loss of circadian PAR bZip transcription factors results in epilepsy
Abstract
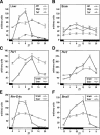
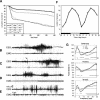
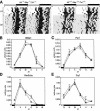
DBP (albumin D-site-binding protein), HLF (hepatic leukemia factor), and TEF (thyrotroph embryonic factor) are the three members of the PAR bZip (proline and acidic amino acid-rich basic leucine zipper) transcription factor family. All three of these transcriptional regulatory proteins accumulate with robust circadian rhythms in tissues with high amplitudes of clock gene expression, such as the suprachiasmatic nucleus (SCN) and the liver. However, they are expressed at nearly invariable levels in most brain regions, in which clock gene expression only cycles with low amplitude. Here we show that mice deficient for all three PAR bZip proteins are highly susceptible to generalized spontaneous and audiogenic epilepsies that frequently are lethal. Transcriptome profiling revealed pyridoxal kinase (Pdxk) as a target gene of PAR bZip proteins in both liver and brain. Pyridoxal kinase converts vitamin B6 derivatives into pyridoxal phosphate (PLP), the coenzyme of many enzymes involved in amino acid and neurotransmitter metabolism. PAR bZip-deficient mice show decreased brain levels of PLP, serotonin, and dopamine, and such changes have previously been reported to cause epilepsies in other systems. Hence, the expression of some clock-controlled genes, such as Pdxk, may have to remain within narrow limits in the brain. This could explain why the circadian oscillator has evolved to generate only low-amplitude cycles in most brain regions.
Figures




References
-
- Battaglioli G., Liu, H., and Martin, D.L. 2003. Kinetic differences between the isoforms of glutamate decarboxylase: Implications for the regulation of GABA synthesis. J. Neurochem. 86: 879-887. - PubMed
-
- Bell R.R. and Haskell, B.E. 1971. Metabolism of vitamin B 6 in the I-strain mouse. I. Absorption, excretion, and conversion of vitamin to enzyme co-factor. Arch. Biochem. Biophys. 147: 588-601. - PubMed
-
- Bell R.R., Blanchard, C.A., and Haskell, B.E. 1971. Metabolism of vitamin B6 in the I-strain mouse. II. Oxidation of pyridoxal. Arch. Biochem. Biophys. 147: 602-611. - PubMed
-
- Blaschko H. and Buffoni, F. 1965. Pyridoxal phosphate as a constituent of the histaminase (benzylamine oxidase) of pig plasma. Proc. R. Soc. Lond. B. Biol. Sci. 163: 45-60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
