Regulation of hypercompetence in Legionella pneumophila
- PMID: 15175295
- PMCID: PMC419971
- DOI: 10.1128/JB.186.12.3814-3825.2004
Regulation of hypercompetence in Legionella pneumophila
Abstract
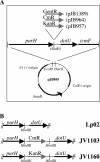
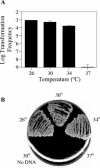
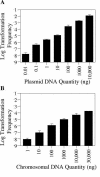
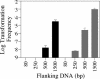
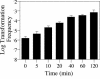
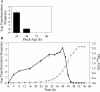
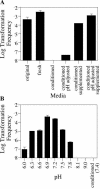
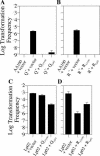
Although many bacteria are known to be naturally competent for DNA uptake, this ability varies dramatically between species and even within a single species, some isolates display high levels of competence while others seem to be completely nontransformable. Surprisingly, many nontransformable bacterial strains appear to encode components necessary for DNA uptake. We believe that many such strains are actually competent but that this ability has been overlooked because standard laboratory conditions are inappropriate for competence induction. For example, most strains of the gram-negative bacterium Legionella pneumophila are not competent under normal laboratory conditions of aerobic growth at 37 degrees C. However, it was previously reported that microaerophilic growth at 37 degrees C allows L. pneumophila serogroup 1 strain AA100 to be naturally transformed. Here we report that another L. pneumophila serogroup 1 strain, Lp02, can also be transformed under these conditions. Moreover, Lp02 can be induced to high levels of competence by a second set of conditions, aerobic growth at 30 degrees C. In contrast to Lp02, AA100 is only minimally transformable at 30 degrees C, indicating that Lp02 is hypercompetent under these conditions. To identify potential causes of hypercompetence, we isolated mutants of AA100 that exhibited enhanced DNA uptake. Characterization of these mutants revealed two genes, proQ and comR, that are involved in regulating competence in L. pneumophila. This approach, involving the isolation of hypercompetent mutants, shows great promise as a method for identifying natural transformation in bacterial species previously thought to be nontransformable.
Figures

References
-
- Behnke, D. 1981. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol. Gen. Genet. 182:490-497. - PubMed
-
- Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

