Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains
- PMID: 15175300
- PMCID: PMC419928
- DOI: 10.1128/JB.186.12.3862-3872.2004
Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains
Abstract
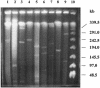
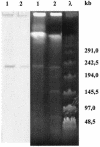
A systematic survey for the presence of plasmids in 17 different xenobiotic-degrading Sphingomonas strains was performed. In almost all analyzed strains, two to five plasmids with sizes of about 50 to 500 kb were detected by using pulsed-field gel electrophoresis. A comparison of plasmid preparations untreated or treated with S1 nuclease suggested that, in general, Sphingomonas plasmids are circular. Hybridization experiments with labeled gene probes suggested that large plasmids are involved in the degradation of dibenzo-p-dioxin, dibenzofuran, and naphthalenesulfonates in S. wittichii RW1, Sphingomonas sp. HH69, and S. xenophaga BN6, respectively. The plasmids which are responsible for the degradation of naphthalene, biphenyl, and toluene by S. aromaticivorans F199 (pNL1) and of naphthalenesulfonates by S. xenophaga BN6 (pBN6) were site-specifically labeled with a kanamycin resistance cassette. The conjugative transfer of these labeled plasmids was attempted with various bacterial strains as putative recipient strains. Thus, a conjugative transfer of plasmid pBN6 from S. xenophaga BN6 to a cured mutant of strain BN6 and to Sphingomonas sp. SS3 was observed. The conjugation experiments with plasmid pNL1 suggested a broader host range of this plasmid, because it was transferred without any obvious structural changes to S. yanoikuyae B1, Sphingomonas sp. SS3, and S. herbicidovorans. In contrast, major plasmid rearrangements were observed in the transconjugants after the transfer of plasmid pNL1 to Sphingomonas sp. HH69 and of pBN6 to Sphingomonas sp. SS3. No indications for the transfer of a Sphingomonas plasmid to bacteria outside of the Sphingomonadaceae were obtained.
Figures



Similar articles
-
The Sphingomonas plasmid pCAR3 is involved in complete mineralization of carbazole.J Bacteriol. 2007 Mar;189(5):2007-20. doi: 10.1128/JB.01486-06. Epub 2006 Dec 15. J Bacteriol. 2007. PMID: 17172338 Free PMC article.
-
Structural and replicative diversity of large plasmids from sphingomonads that degrade polycyclic aromatic compounds and xenobiotics.Microbiology (Reading). 2005 Jun;151(Pt 6):2025-2037. doi: 10.1099/mic.0.27965-0. Microbiology (Reading). 2005. PMID: 15942009
-
Identification and functional analysis of the genes for naphthalenesulfonate catabolism by Sphingomonas xenophaga BN6.Microbiology (Reading). 2006 Jul;152(Pt 7):1929-1940. doi: 10.1099/mic.0.28783-0. Microbiology (Reading). 2006. PMID: 16804169
-
Molecular characteristics of xenobiotic-degrading sphingomonads.Appl Microbiol Biotechnol. 2009 Jan;81(5):793-811. doi: 10.1007/s00253-008-1752-3. Epub 2008 Nov 11. Appl Microbiol Biotechnol. 2009. PMID: 19002456 Review.
-
The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs).J Gen Appl Microbiol. 2003 Feb;49(1):1-19. doi: 10.2323/jgam.49.1. J Gen Appl Microbiol. 2003. PMID: 12682862 Review.
Cited by
-
Impact of microbes on autoimmune diseases.Arch Immunol Ther Exp (Warsz). 2013 Jun;61(3):175-86. doi: 10.1007/s00005-013-0216-3. Epub 2013 Feb 16. Arch Immunol Ther Exp (Warsz). 2013. PMID: 23417246 Free PMC article. Review.
-
The variations of native plasmids greatly affect the cell surface hydrophobicity of sphingomonads.mSystems. 2023 Dec 21;8(6):e0086223. doi: 10.1128/msystems.00862-23. Epub 2023 Nov 1. mSystems. 2023. PMID: 37909742 Free PMC article.
-
Biochemistry of microbial degradation of hexachlorocyclohexane and prospects for bioremediation.Microbiol Mol Biol Rev. 2010 Mar;74(1):58-80. doi: 10.1128/MMBR.00029-09. Microbiol Mol Biol Rev. 2010. PMID: 20197499 Free PMC article. Review.
-
The Sphingomonas plasmid pCAR3 is involved in complete mineralization of carbazole.J Bacteriol. 2007 Mar;189(5):2007-20. doi: 10.1128/JB.01486-06. Epub 2006 Dec 15. J Bacteriol. 2007. PMID: 17172338 Free PMC article.
-
Comparative metatranscriptome analysis revealed broad response of microbial communities in two soil types, agriculture versus organic soil.J Genet Eng Biotechnol. 2019 Oct 14;17(1):6. doi: 10.1186/s43141-019-0006-3. J Genet Eng Biotechnol. 2019. PMID: 31659568 Free PMC article.
References
-
- Balkwill, D. L., G. R. Drake, R. H. Reeves, J. K. Fredrickson, D. C. White, D. B. Ringelberg, D. P. Chandler, M. F. Romine, D. W. Kennedy, and C. M. Spadoni. 1997. Taxonomic study of aromatic-degrading bacteria from deep-terrestrial-subsurface sediments and description of Sphingomonas aromaticivorans sp. nov., Sphingomonas subterranea sp. nov., and Sphingomonas stygia sp. nov. Int. J. Syst. Bacteriol. 47:191-201. - PubMed
-
- Baraniecki, C. A., J. Aislabie, and J. M. Foght. 2002. Characterization of Sphingomonas sp. Ant 17, an aromatic hydrocarbon-degrading bacterium isolated from Antarctic soil. Microb. Ecol. 43:44-54. - PubMed
-
- Barton, B. M., G. P. Hardening, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:234-240. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

