Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK
- PMID: 15175315
- PMCID: PMC419961
- DOI: 10.1128/JB.186.12.4014-4018.2004
Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK
Abstract
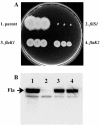
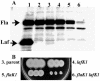
Gene organization and hierarchical regulation of the polar flagellar genes of Vibrio parahaemolyticus, Vibrio cholerae, and Pseudomonas aeruginosa appear highly similar, with one puzzling difference. Two sigma(54)-dependent regulators are required to direct different classes of intermediate flagellar gene expression in V. cholerae and P. aeruginosa, whereas the V. parahaemolyticus homolog of one of these regulators, FlaK, appears dispensable. Here we demonstrate that there is compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK.
Figures


Similar articles
-
Lateral flagellar gene system of Vibrio parahaemolyticus.J Bacteriol. 2003 Aug;185(15):4508-18. doi: 10.1128/JB.185.15.4508-4518.2003. J Bacteriol. 2003. PMID: 12867460 Free PMC article.
-
Deciphering bacterial flagellar gene regulatory networks in the genomic era.Adv Appl Microbiol. 2009;67:257-95. doi: 10.1016/S0065-2164(08)01008-3. Adv Appl Microbiol. 2009. PMID: 19245942 Review.
-
Dual flagellar systems enable motility under different circumstances.J Mol Microbiol Biotechnol. 2004;7(1-2):18-29. doi: 10.1159/000077866. J Mol Microbiol Biotechnol. 2004. PMID: 15170400 Review.
-
Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of a flagellin.Mol Microbiol. 1996 Apr;20(1):137-49. doi: 10.1111/j.1365-2958.1996.tb02496.x. Mol Microbiol. 1996. PMID: 8861212
-
The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae.Mol Microbiol. 2001 Mar;39(6):1595-609. doi: 10.1046/j.1365-2958.2001.02348.x. Mol Microbiol. 2001. PMID: 11260476
Cited by
-
Transcriptome Analysis Reveals Cross-Talk between the Flagellar Transcriptional Hierarchy and Secretion System in Plesiomonas shigelloides.Int J Mol Sci. 2024 Jul 5;25(13):7375. doi: 10.3390/ijms25137375. Int J Mol Sci. 2024. PMID: 39000482 Free PMC article.
-
Environment-directed activation of the Escherichia coliflhDC operon by transposons.Microbiology (Reading). 2017 Apr;163(4):554-569. doi: 10.1099/mic.0.000426. Epub 2017 Apr 12. Microbiology (Reading). 2017. PMID: 28100305 Free PMC article.
-
Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes.J Bacteriol. 2011 Oct;193(19):5179-90. doi: 10.1128/JB.05355-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784933 Free PMC article.
-
Characterization of the relationship between polar and lateral flagellar structural genes in the deep-sea bacterium Shewanella piezotolerans WP3.Sci Rep. 2016 Dec 22;6:39758. doi: 10.1038/srep39758. Sci Rep. 2016. PMID: 28004809 Free PMC article.
-
Characterizing the Adherence Profiles of Virulent Vibrio parahaemolyticus Isolates.Microb Ecol. 2018 Jan;75(1):152-162. doi: 10.1007/s00248-017-1025-8. Epub 2017 Jul 17. Microb Ecol. 2018. PMID: 28717834
References
-
- Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. - PubMed
-
- Correa, N. E., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. - PubMed
-
- Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

