Construction of oligonucleotide arrays on a glass surface using a heterobifunctional reagent, N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilylpropyl-3-amine (NTMTA)
- PMID: 15175428
- PMCID: PMC434455
- DOI: 10.1093/nar/gnh075
Construction of oligonucleotide arrays on a glass surface using a heterobifunctional reagent, N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilylpropyl-3-amine (NTMTA)
Abstract
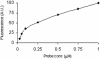
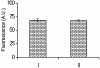
A rapid method for construction of oligonucleotide arrays on a glass surface, using a novel heterobifunctional reagent, N-(2-trifluoroethanesulfonatoethyl)-N-(methyl)-triethoxysilylpropyl-3-amine (NTMTA), has been described. The heterobifunctional reagent, NTMTA, carries two different thermoreactive groups. The triethoxysilyl group on one end is specific towards silanol functions on the virgin glass surface, while the trifluoroethanesulfonyl (tresyl) group on the other end of the reagent reacts specifically with aminoalkyl- or mercaptoalkyl- functionalized oligonucleotides. Immobilization of oligonucleotides on a glass surface has been realized via two routes. In the first one (A), 5'- aminoalkyl- or mercaptoalkyl-functionalized oligonucleotides were allowed to react with NTMTA to form a oligonucleotide-triethoxysilyl conjugate which, in a subsequent reaction with unmodified (virgin) glass microslide, results in surface-bound oligonucleotides. In the second route (B), the NTMTA reagent reacts first with a glass microslide whereby it generates trifluoroethanesulfonate ester functions on it, which in a subsequent step react with 5'-aminoalkyl or mercaptoalkyl oligonucleotides to generate support-bound oligonucleotides. Subsequently, the oligonucleotide arrays prepared by both routes were analyzed by hybridization experiments with complementary oligonucleotides. The constructed microarrays were successfully used in single and multiple nucleotide mismatch detection by hybridizing these with fluorescein-labeled complementary oligonucleotides. Further more, the proposed method was compared with the existing methods with respect to immobilization efficiency of oligonucleotides.
Figures











Similar articles
-
A new heterobifunctional reagent for immobilization of biomolecules on glass surface.Bioorg Med Chem Lett. 2004 Mar 8;14(5):1097-9. doi: 10.1016/j.bmcl.2003.12.077. Bioorg Med Chem Lett. 2004. PMID: 14980643
-
Construction of oligonucleotide microarrays (biochip) using heterobifunctional reagents.Methods Mol Biol. 2007;381:133-63. doi: 10.1007/978-1-59745-303-5_7. Methods Mol Biol. 2007. PMID: 17984518
-
N-(3-Triethoxysilylpropyl)-6-(N-maleimido)-hexanamide: An efficient heterobifunctional reagent for the construction of oligonucleotide microarrays.Anal Biochem. 2006 Oct 15;357(2):240-8. doi: 10.1016/j.ab.2006.07.006. Epub 2006 Aug 4. Anal Biochem. 2006. PMID: 16930520
-
Strategies in the preparation of DNA oligonucleotide arrays for diagnostic applications.Curr Med Chem. 2001 Aug;8(10):1213-44. doi: 10.2174/0929867013372463. Curr Med Chem. 2001. PMID: 11472237 Review.
-
Arrays of immobilized oligonucleotides--contributions to nucleic acids technology.Curr Pharm Biotechnol. 2003 Dec;4(6):379-95. doi: 10.2174/1389201033377454. Curr Pharm Biotechnol. 2003. PMID: 14683432 Review.
Cited by
-
Controlled loading of oligodeoxyribonucleotide monolayers onto unoxidized crystalline silicon; fluorescence-based determination of the surface coverage and of the hybridization efficiency; parallel imaging of the process by Atomic Force Microscopy.Nucleic Acids Res. 2006 Feb 28;34(4):e32. doi: 10.1093/nar/gnj034. Nucleic Acids Res. 2006. PMID: 16507670 Free PMC article.
-
Waveguide-mode sensors as aptasensors.Sensors (Basel). 2012;12(2):2136-51. doi: 10.3390/s120202136. Epub 2012 Feb 15. Sensors (Basel). 2012. PMID: 22438756 Free PMC article. Review.
References
-
- Ramsay G. (1998) DNA-chips: state-of-the-art. Nat. Biotechnol., 16, 40–44. - PubMed
-
- Pollock J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A. Williams,C.F., Jeffery S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DNA-copy number changes using cDNA microarrays. Nature Genet., 23, 41–46. - PubMed
-
- Marshall A. and Hodgson,J. (1998) DNA chips: an array of possibilities. Nat. Biotechnol., 16, 27–31. - PubMed
-
- Drobyshev A.N., Mologina,N., Shick,V., Pobedimskaya D., Yershov,G. and Mirzabekov,A. (1997) Sequence analysis by hybridization with oligonucleotide microchip: identification of β-thalessemia mutations. Gene, 188, 45–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

