A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing
- PMID: 15175653
- PMCID: PMC423283
- DOI: 10.1038/sj.emboj.7600241
A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing
Abstract
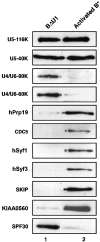
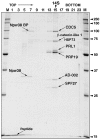
During catalytic activation of the spliceosome, snRNP remodeling events occur, leading to the formation of a 35S U5 snRNP that contains a large group of proteins, including Prp19 and CDC5, not found in 20S U5 snRNPs. To investigate the function of 35S U5 proteins, we immunoaffinity purified human spliceosomes that had not yet undergone catalytic activation (designated BDeltaU1), which contained U2, U4, U5, and U6, but lacked U1 snRNA. Comparison of the protein compositions of BDeltaU1 and activated B* spliceosomes revealed that, whereas U4/U6 snRNP proteins are stably associated with BDeltaU1 spliceosomes, 35S U5-associated proteins (which are present in B*) are largely absent, suggesting that they are dispensable for complex B formation. Indeed, immunodepletion/complementation experiments demonstrated that a subset of 35S U5 proteins including Prp19, which form a stable heteromeric complex, are required prior to catalytic step 1 of splicing, but not for stable integration of U4/U6.U5 tri-snRNPs. Thus, comparison of the proteomes of spliceosomal complexes at defined stages can provide information as to which proteins function as a group at a particular step of splicing.
Figures
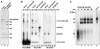


References
-
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 - PubMed
-
- Burge CB, Tuschl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland C, Atkins (eds), 2nd edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press pp 525–560
-
- Chan SP, Kao DI, Tsai WY, Cheng SC (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

