PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits
- PMID: 15175657
- PMCID: PMC449775
- DOI: 10.1038/sj.emboj.7600265
PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits
Abstract
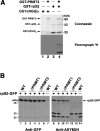
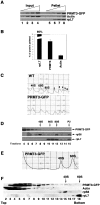
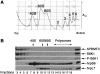
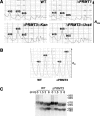
The mammalian protein arginine methyltransferase 3 (PRMT3) catalyzes the formation of asymmetric (type I) dimethylarginine in vitro. As yet, natural substrates and cellular pathways modulated by PRMT3 remain unknown. Here, we have identified an ortholog of PRMT3 in fission yeast. Tandem affinity purification of fission yeast PRMT3 coupled with mass spectrometric protein identification revealed that PRMT3 associates with components of the translational machinery. We identified the 40S ribosomal protein S2 as the first physiological substrate of PRMT3. In addition, a fraction of yeast and human PRMT3 cosedimented with free 40S ribosomal subunits, as determined by sucrose gradient velocity centrifugation. The activity of PRMT3 is not essential since prmt3-disrupted cells are viable. Interestingly, cells lacking PRMT3 showed an accumulation of free 60S ribosomal subunits resulting in an imbalance in the 40S:60S free subunits ratio; yet pre-rRNA processing appeared to occur normally. Our results identify PRMT3 as the first type I ribosomal protein arginine methyltransferase and suggest that it regulates ribosome biosynthesis at a stage beyond pre-rRNA processing.
Figures



References
-
- Akiyoshi Y, Clayton J, Phan L, Yamamoto M, Hinnebusch AG, Watanabe Y, Asano K (2001) Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem 276: 10056–10062 - PubMed
-
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S (2000) Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem 275: 16030–16036 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

