How many novel eukaryotic 'kingdoms'? Pitfalls and limitations of environmental DNA surveys
- PMID: 15176975
- PMCID: PMC428588
- DOI: 10.1186/1741-7007-2-13
How many novel eukaryotic 'kingdoms'? Pitfalls and limitations of environmental DNA surveys
Abstract
Background: Over the past few years, the use of molecular techniques to detect cultivation-independent, eukaryotic diversity has proven to be a powerful approach. Based on small-subunit ribosomal RNA (SSU rRNA) gene analyses, these studies have revealed the existence of an unexpected variety of new phylotypes. Some of them represent novel diversity in known eukaryotic groups, mainly stramenopiles and alveolates. Others do not seem to be related to any molecularly described lineage, and have been proposed to represent novel eukaryotic kingdoms. In order to review the evolutionary importance of this novel high-level eukaryotic diversity critically, and to test the potential technical and analytical pitfalls and limitations of eukaryotic environmental DNA surveys (EES), we analysed 484 environmental SSU rRNA gene sequences, including 81 new sequences from sediments of the small river, the Seymaz (Geneva, Switzerland).
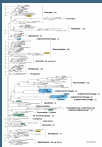
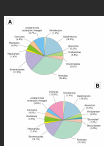
Results: Based on a detailed screening of an exhaustive alignment of eukaryotic SSU rRNA gene sequences and the phylogenetic re-analysis of previously published environmental sequences using Bayesian methods, our results suggest that the number of novel higher-level taxa revealed by previously published EES was overestimated. Three main sources of errors are responsible for this situation: (1) the presence of undetected chimeric sequences; (2) the misplacement of several fast-evolving sequences; and (3) the incomplete sampling of described, but yet unsequenced eukaryotes. Additionally, EES give a biased view of the diversity present in a given biotope because of the difficult amplification of SSU rRNA genes in some taxonomic groups.
Conclusions: Environmental DNA surveys undoubtedly contribute to reveal many novel eukaryotic lineages, but there is no clear evidence for a spectacular increase of the diversity at the kingdom level. After re-analysis of previously published data, we found only five candidate lineages of possible novel high-level eukaryotic taxa, two of which comprise several phylotypes that were found independently in different studies. To ascertain their taxonomic status, however, the organisms themselves have now to be identified.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

