Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping
- PMID: 1518053
- PMCID: PMC2899704
- DOI: 10.1016/0022-2836(92)91063-u
Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping
Abstract
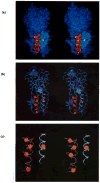
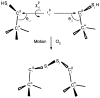
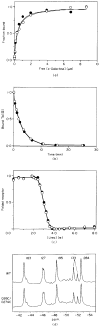
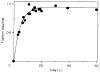
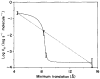
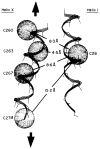
The D-galactose chemosensory receptor of Escherichia coli is a .32 kDa globular protein possessing two distinct structural domains, each organized in an alpha/beta folding motif. Helices I and X lie at adjacent approximately parallel positions on the surface of the N-terminal domain, near the hinge region. In order to analyze the relative thermal motions of these two helices, the present study utilizes a generalizable disulfide trapping approach: first, site-directed mutagenesis is used to place a pair of cysteine residues at locations of interest on the protein surface, then disulfide bond formation is used to trap intramolecular cysteine-cysteine collisions resulting from thermal motions. Specifically, four engineered di-cysteine receptors have been constructed, each possessing one cysteine at position 26 on helix I, and a second cysteine at varying positions on helix X. A fifth control receptor possesses one cysteine at position 26, and a second on the opposite surface of the molecule. These surface cysteine substitutions have little or no effect on the measurable receptor parameters as judged by ligand binding equilibria and kinetics, protein stability, and 19F nuclear magnetic resonance, indicating that the engineered receptors are useful probes of native backbone dynamics. Spatial and kinetic features of backbone motions have been investigated by measuring intramolecular disulfide formation rates for cysteine pairs in the fully liganded receptor. The resulting rates decrease monotonically with increasing distance between cysteines in the crystal structure, while no disulfide formation is observed for the control pair unless the molecule is unfolded. The minimum translational amplitudes of the observed backbone motions range from 4.5 to 15.2 A, and the minimum rotational amplitudes are as large as 35 degrees. For each motion the rate of intramolecular sulfhydryl-sulfhydryl collision has been estimated from the measured rate of disulfide formation: the 4.5 and 15.2 A translations yield approximately 10(4) and approximately 10 collisions s-1 molecule-1, respectively. These collision rates, which are faster than ligand dissociation, likely underestimate the actual motional frequencies since only an undetermined fraction of the total motions yield collisions. The simplest plausible trajectory capable of producing such collisions is a rate-limiting translation of one or both helices along their long axes, coupled with minor helix rotations. When sugar is removed from the receptor, a substantial increase in backbone dynamics is observed, indicating the presence of new long-range backbone trajectories. Overall, the results suggest that internal motions in proteins may have larger amplitudes than previously observed.
Figures
References
-
- Altenbach C, Marti T, Khorana HG, Hubbell WL. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990;248:1088–1092. - PubMed
-
- Argos P, Mahoney WC, Hermodson MA, Hanei M. Structural prediction of sugar-binding proteins functional in chemotaxis and transport. J Biol Chem. 1981;256:4357–4361. - PubMed
-
- Balaji VN, Mobasser A, Rao SN. Modification of protein stability by introduction of disulfide bridges and prolines: geometric criteria for mutation sites. Biochem Biophys Res Commun. 1989;160:109–114. - PubMed
-
- Boos W, Gordon AS, Hall RE, Price DH. Transport properties of the galactose-binding protein of Escherichia coli: substrate induced conformational change. J Biol Chem. 1972;247:917–924. - PubMed
-
- Bowie JU, Reidhaarolson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990;247:1306–1310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

