LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1
- PMID: 15181175
- PMCID: PMC434442
- DOI: 10.1093/nar/gkh636
LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1
Erratum in
- Nucleic Acids Res. 2004 Jun 29;32(11):3512
Abstract
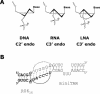
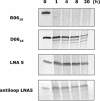
One of the major limitations of the use of phosphodiester oligonucleotides in cells is their rapid degradation by nucleases. To date, several chemical modifications have been employed to overcome this issue but insufficient efficacy and/or specificity have limited their in vivo usefulness. In this work conformationally restricted nucleotides, locked nucleic acid (LNA), were investigated to design nuclease resistant aptamers targeted against the HIV-1 TAR RNA. LNA/DNA chimeras were synthesized from a shortened version of the hairpin RNA aptamer identified by in vitro selection against TAR. The results indicate that these modifications confer good protection towards nuclease digestion. Electrophoretic mobility shift assays, thermal denaturation monitored by UV-spectroscopy and surface plasmon resonance experiments identified LNA/DNA TAR ligands that bind to TAR with a dissociation constant in the low nanomolar range as the parent RNA aptamer. The crucial G, A residues that close the aptamer loop remain a key structural determinant for stable LNA/DNA chimera-TAR complexes. This work provides evidence that LNA modifications alternated with DNA can generate stable structured RNA mimics for interacting with folded RNA targets.
Figures

References
-
- Berkhout B. (2000) Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol., 48, 29–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

