AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis
- PMID: 15181201
- PMCID: PMC514136
- DOI: 10.1104/pp.104.044495
AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis
Abstract
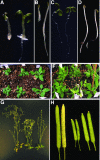
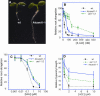
Auxin affects many aspects of plant growth and development. We previously used chemical genetics to dissect auxin-signaling mechanisms and identified a small molecule, sirtinol, that constitutively activated auxin signaling (Y. Zhao et al. [2003], Science 301: 1107-1110). Here we describe the isolation, characterization, and cloning of an Arabidopsis mutant Atcand1-1 that emerged from a genetic screen for mutants insensitive to sirtinol. Loss-of-function mutants of AtCAND1 were resistant to sirtinol and auxin, but not to gibberellins or brassinolide. Atcand1 displayed developmental phenotypes similar to those of axr1, namely, short petioles, downwardly curling leaves, short inflorescence, and reduced fertility. AtCAND1 is homologous to human CAND1, a protein that is composed almost entirely of HEAT-repeat units and has been implicated in regulating the assembly and disassembly of the SCF protein degradation machinery. Taken together with previous biochemical studies, this work helps to elucidate the roles of AtCAND1 in protein degradation and auxin signaling.
Figures


References
-
- Ballas N, Wong LM, Theologis A (1993) Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J Mol Biol 233: 580–596 - PubMed
-
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 - PubMed
-
- Cope GA, Deshaies RJ (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

