Intracellular localization of Arabidopsis sulfurtransferases
- PMID: 15181206
- PMCID: PMC514126
- DOI: 10.1104/pp.104.040121
Intracellular localization of Arabidopsis sulfurtransferases
Abstract
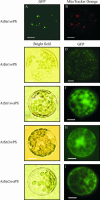
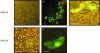
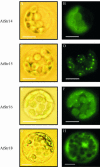
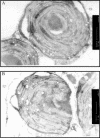
Sulfurtransferases (Str) comprise a group of enzymes widely distributed in archaea, eubacteria, and eukaryota which catalyze the transfer of a sulfur atom from suitable sulfur donors to nucleophilic sulfur acceptors. In all organisms analyzed to date, small gene families encoding Str proteins have been identified. The gene products were localized to different compartments of the cells. Our interest concerns the localization of Str proteins encoded in the nuclear genome of Arabidopsis. Computer-based prediction methods revealed localization in different compartments of the cell for six putative AtStrs. Several methods were used to determine the localization of the AtStr proteins experimentally. For AtStr1, a mitochondrial localization was demonstrated by immunodetection in the proteome of isolated mitochondria resolved by one- and two-dimensional gel electrophoresis and subsequent blotting. The respective mature AtStr1 protein was identified by mass spectrometry sequencing. The same result was obtained by transient expression of fusion constructs with the green fluorescent protein in Arabidopsis protoplasts, whereas AtStr2 was exclusively localized to the cytoplasm by this method. Three members of the single-domain AtStr were localized in the chloroplasts as demonstrated by transient expression of green fluorescent protein fusions in protoplasts and stomata, whereas the single-domain AtStr18 was shown to be cytoplasmic. The remarkable subcellular distribution of AtStr15 was additionally analyzed by transmission electron immunomicroscopy using a monospecific antibody against green fluorescent protein, indicating an attachment to the thylakoid membrane. The knowledge of the intracellular localization of the members of this multiprotein family will help elucidate their specific functions in the organism.
Figures


Similar articles
-
Characterization of the sulfurtransferase family from Oryza sativa L.Plant Physiol Biochem. 2011 Sep;49(9):1064-70. doi: 10.1016/j.plaphy.2011.07.010. Epub 2011 Jul 23. Plant Physiol Biochem. 2011. PMID: 21821426
-
Differential expression of Arabidopsis sulfurtransferases under various growth conditions.Plant Physiol Biochem. 2007 Mar-Apr;45(3-4):178-87. doi: 10.1016/j.plaphy.2007.02.005. Epub 2007 Feb 25. Plant Physiol Biochem. 2007. PMID: 17408957
-
Plant mercaptopyruvate sulfurtransferases: molecular cloning, subcellular localization and enzymatic activities.Eur J Biochem. 2000 Sep;267(17):5621-30. doi: 10.1046/j.1432-1327.2000.01633.x. Eur J Biochem. 2000. PMID: 10951223
-
Latest news about the sulfurtransferase protein family of higher plants.Amino Acids. 2011 Jun;41(1):43-57. doi: 10.1007/s00726-010-0478-6. Epub 2010 Feb 5. Amino Acids. 2011. PMID: 20135153 Review.
-
Sulfotransferase pharmacogenetics.Pharmacol Ther. 1990;45(1):93-107. doi: 10.1016/0163-7258(90)90010-y. Pharmacol Ther. 1990. PMID: 2405435 Review. No abstract available.
Cited by
-
Cellular and Subcellular Immunohistochemical Localization and Quantification of Cadmium Ions in Wheat (Triticum aestivum).PLoS One. 2015 May 5;10(5):e0123779. doi: 10.1371/journal.pone.0123779. eCollection 2015. PLoS One. 2015. PMID: 25941807 Free PMC article.
-
Organelle Interactions in Plant Cells.Results Probl Cell Differ. 2024;73:43-69. doi: 10.1007/978-3-031-62036-2_3. Results Probl Cell Differ. 2024. PMID: 39242374 Review.
-
Solution structure of a single-domain thiosulfate sulfurtransferase from Arabidopsis thaliana.Protein Sci. 2006 Dec;15(12):2836-41. doi: 10.1110/ps.062395206. Epub 2006 Nov 6. Protein Sci. 2006. PMID: 17088324 Free PMC article.
-
GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis.Plant Cell. 2009 Apr;21(4):1109-28. doi: 10.1105/tpc.108.065250. Epub 2009 Apr 17. Plant Cell. 2009. PMID: 19376934 Free PMC article.
-
Mitochondrial biogenesis and function in Arabidopsis.Arabidopsis Book. 2008;6:e0111. doi: 10.1199/tab.0111. Epub 2008 Jul 9. Arabidopsis Book. 2008. PMID: 22303236 Free PMC article.
References
-
- Abel S, Theologis A (1998) Transient gene expression in protoplasts of Arabidopsis thaliana. Methods Mol Biol 82: 209–217 - PubMed
-
- Bauer M, Papenbrock J (2002) Identification and characterization of single-domain thiosulfate sulfurtransferases from Arabidopsis thaliana. FEBS Lett 532: 427–431 - PubMed
-
- Bonomi F, Pagani S, Cerletti P, Cannella C (1977) Rhodanese-mediated sulfur transfer to succinate dehydrogenase. Eur J Biochem 72: 17–24 - PubMed
-
- Cerletti P (1986) Seeking a better job for an under-employed enzyme: rhodanese. Trends Biochem Sci 11: 369–372
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

