Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein
- PMID: 15184046
- PMCID: PMC7111000
- DOI: 10.1016/j.bbrc.2004.05.046
Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein
Abstract
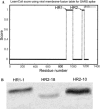
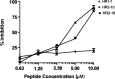
Heptad repeat regions (HR1 and HR2) are highly conserved sequences located in the glycoproteins of enveloped viruses. They form a six-helix bundle structure and are important in the process of virus fusion. Peptides derived from the HR regions of some viruses have been shown to inhibit the entry of these viruses. SARS-CoV was also predicted to have HR1 and HR2 regions in the S2 protein. Based on this prediction, we designed 25 peptides and screened them using a HIV-luc/SARS pseudotyped virus assay. Two peptides, HR1-1 and HR2-18, were identified as potential inhibitors, with EC(50) values of 0.14 and 1.19microM, respectively. The inhibitory effects of these peptides were validated by the wild-type SARS-CoV assay. HR1-1 and HR2-18 can serve as functional probes for dissecting the fusion mechanism of SARS-CoV and also provide the potential of further identifying potent inhibitors for SARS-CoV entry.
Figures

Similar articles
-
Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8455-60. doi: 10.1073/pnas.0400576101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150417 Free PMC article.
-
Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors.Lancet. 2004 Mar 20;363(9413):938-47. doi: 10.1016/S0140-6736(04)15788-7. Lancet. 2004. PMID: 15043961 Free PMC article.
-
Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555
-
Multimerization of the heptad repeat regions of the SARS-CoV 2 spike protein.Biochim Biophys Acta Biomembr. 2024 Feb;1866(2):184259. doi: 10.1016/j.bbamem.2023.184259. Epub 2023 Dec 5. Biochim Biophys Acta Biomembr. 2024. PMID: 38061554 Review.
-
Severe acute respiratory syndrome coronavirus entry into host cells: Opportunities for therapeutic intervention.Med Res Rev. 2006 Jul;26(4):414-33. doi: 10.1002/med.20055. Med Res Rev. 2006. PMID: 16521129 Free PMC article. Review.
Cited by
-
Amino acids 1055 to 1192 in the S2 region of severe acute respiratory syndrome coronavirus S protein induce neutralizing antibodies: implications for the development of vaccines and antiviral agents.J Virol. 2005 Mar;79(6):3289-96. doi: 10.1128/JVI.79.6.3289-3296.2005. J Virol. 2005. PMID: 15731223 Free PMC article.
-
Characterization of neutralizing monoclonal antibodies recognizing a 15-residues epitope on the spike protein HR2 region of severe acute respiratory syndrome coronavirus (SARS-CoV).J Biomed Sci. 2005 Oct;12(5):711-27. doi: 10.1007/s11373-005-9004-3. Epub 2005 Nov 9. J Biomed Sci. 2005. PMID: 16132115 Free PMC article.
-
Cannabinoids Block Cellular Entry of SARS-CoV-2 and the Emerging Variants.J Nat Prod. 2022 Jan 28;85(1):176-184. doi: 10.1021/acs.jnatprod.1c00946. Epub 2022 Jan 10. J Nat Prod. 2022. PMID: 35007072 Free PMC article.
-
Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains.Nat Commun. 2017 Apr 10;8:15092. doi: 10.1038/ncomms15092. Nat Commun. 2017. PMID: 28393837 Free PMC article.
-
Comprehensive review on the prevailing COVID-19 therapeutics and the potential of repurposing SARS-CoV-1 candidate drugs to target SARS-CoV-2 as a fast-track treatment and prevention option.Ann Transl Med. 2020 Oct;8(19):1247. doi: 10.21037/atm-20-4071. Ann Transl Med. 2020. PMID: 33178779 Free PMC article. Review.
References
-
- Tsang K.W, Ho P.L, Ooi G.C, Yee W.K, Wang T, Chan-Yeung M, Lam W.K, Seto W.H, Yam L.Y, Cheung T.M, Wong P.C, Lam B, Ip M.S, Chan J, Yuen K.Y, Lai K.N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1977–1985. - PubMed
-
- Poutanen S.M, Low D.E, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M, Chan A.K, Skowronski D.M, Salit I, Simor A.E, Slutsky A.S, Doyle P.W, Krajden M, Petric M, Brunham R.C, McGeer A.J. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348:1995–2005. - PubMed
-
- Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt H.R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier R.A, Berger A, Burguiere A.M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J.C, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk H.D, Osterhaus A.D, Schmitz H, Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

