Enzyme production-based approach for determining the functions of microorganisms within a community
- PMID: 15184128
- PMCID: PMC427761
- DOI: 10.1128/AEM.70.6.3329-3337.2004
Enzyme production-based approach for determining the functions of microorganisms within a community
Abstract
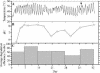
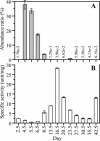
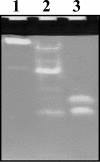
The functions of specific microorganisms in a microbial community were investigated during the composting process. Cerasibacillus quisquiliarum strain BLx(T) and Bacillus thermoamylovorans strain BTa were isolated and characterized in our previous studies based on their dominance in the composting system. Strain BLx(T) degrades gelatin, while strain BTa degrades starch. We hypothesized that these strains play roles in gelatinase and amylase production, respectively. The relationship between changes in the abundance ratios of each strain and those of each enzyme activity during the composting process was examined to address this hypothesis. The increase in gelatinase activity in the compost followed a dramatic increase in the abundance ratio of strain BLx(T). Zymograph analysis demonstrated that the pattern of active gelatinase bands from strain BLx(T) was similar to that from the compost. Gelatinases from both BLx(T) and compost were partially purified and compared. Homologous N-terminal amino acid sequences were found in one of the gelatinases from strain BLx(T) and that of compost. These results indicate strain BLx(T) produces gelatinases during the composting process. Meanwhile, the increase in the abundance ratio of strain BTa was not concurrent with that of amylase activity in the compost. Moreover, the amylase activity pattern of strain BTa on the zymogram was different from that of the compost sample. These results imply that strain BTa may not produce amylases during the composting process. To our knowledge, this is the first report demonstrating that the function of a specific microorganism is directly linked to a function in the community, as determined by culture-independent and enzyme-level approaches.
Figures



Similar articles
-
Bacterial community in the personal-use composting reactor revealed by isolation and cultivation-independent method.J Environ Sci Health B. 2010 Jul;45(5):372-8. doi: 10.1080/03601231003799895. J Environ Sci Health B. 2010. PMID: 20512727
-
[Purification and characterization of thermostable amylases from two bacterial species].Wei Sheng Wu Xue Bao. 2008 Feb;48(2):169-75. Wei Sheng Wu Xue Bao. 2008. PMID: 18437997 Chinese.
-
Inoculation of Pichia kudriavzevii RB1 degrades the organic acids present in raw compost material and accelerates composting.Bioresour Technol. 2013 Sep;144:521-8. doi: 10.1016/j.biortech.2013.07.005. Epub 2013 Jul 8. Bioresour Technol. 2013. PMID: 23886646
-
Application of compost for effective bioremediation of organic contaminants and pollutants in soil.Appl Microbiol Biotechnol. 2016 Apr;100(8):3433-49. doi: 10.1007/s00253-016-7378-y. Epub 2016 Feb 27. Appl Microbiol Biotechnol. 2016. PMID: 26921182 Review.
-
Inactivation of Salmonella Senftenberg strain W 775 during composting of biowastes and garden wastes.J Appl Microbiol. 2007 Jul;103(1):53-64. doi: 10.1111/j.1365-2672.2006.03224.x. J Appl Microbiol. 2007. PMID: 17584452 Review.
Cited by
-
Microbial community structure and dynamics of dark fire-cured tobacco fermentation.Appl Environ Microbiol. 2007 Feb;73(3):825-37. doi: 10.1128/AEM.02378-06. Epub 2006 Dec 1. Appl Environ Microbiol. 2007. PMID: 17142368 Free PMC article.
-
Changes in bacterial communities accompanied by aggregation in a fed-batch composting reactor.Curr Microbiol. 2008 May;56(5):458-67. doi: 10.1007/s00284-008-9107-y. Epub 2008 Jan 30. Curr Microbiol. 2008. PMID: 18231830
References
-
- Beffa, T., M. Blanc, L. Marilley, J. L. Fischer, P. F. Lyon, and M. Aragno. 1996. Taxonomic and metabolic microbial diversity during composting, p. 149-161. In M. de Bertoldi, P. Sequi, B. Lemmes, and T. Papi (ed.), The science of composting, part I. Chapman and Hall, London, United Kingdom.
-
- Caraway, W. T. 1959. A stable starch substrate for the determination of amylase in serum and other body fluids. Am. J. Clin. Pathol. 32:97-99. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

