Biotransformation of explosives by the old yellow enzyme family of flavoproteins
- PMID: 15184158
- PMCID: PMC427764
- DOI: 10.1128/AEM.70.6.3566-3574.2004
Biotransformation of explosives by the old yellow enzyme family of flavoproteins
Abstract
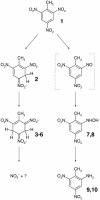
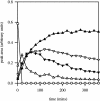
Several independent studies of bacterial degradation of nitrate ester explosives have demonstrated the involvement of flavin-dependent oxidoreductases related to the old yellow enzyme (OYE) of yeast. Some of these enzymes also transform the nitroaromatic explosive 2,4,6-trinitrotoluene (TNT). In this work, catalytic capabilities of five members of the OYE family were compared, with a view to correlating structure and function. The activity profiles of the five enzymes differed substantially; no one compound proved to be a good substrate for all five enzymes. TNT is reduced, albeit slowly, by all five enzymes. The nature of the transformation products differed, with three of the five enzymes yielding products indicative of reduction of the aromatic ring. Our findings suggest two distinct pathways of TNT transformation, with the initial reduction of TNT being the key point of difference between the enzymes. Characterization of an active site mutant of one of the enzymes suggests a structural basis for this difference.
Figures



Similar articles
-
Insights into the biotransformation of 2,4,6-trinitrotoluene by the old yellow enzyme family of flavoproteins. A computational study.Phys Chem Chem Phys. 2019 Jun 5;21(22):11589-11598. doi: 10.1039/c8cp07873d. Phys Chem Chem Phys. 2019. PMID: 30801593
-
Degradation of explosives by nitrate ester reductases.Biochem Soc Symp. 2001;(68):143-53. doi: 10.1042/bss0680143. Biochem Soc Symp. 2001. PMID: 11573344
-
Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase.Appl Environ Microbiol. 1998 Aug;64(8):2864-8. doi: 10.1128/AEM.64.8.2864-2868.1998. Appl Environ Microbiol. 1998. PMID: 9687442 Free PMC article.
-
Degradation of nitrate ester and nitroaromatic explosives by Enterobacter cloacae PB2.Biochem Soc Trans. 1998 Nov;26(4):680-5. doi: 10.1042/bst0260680. Biochem Soc Trans. 1998. PMID: 10047806 Review. No abstract available.
-
Microbial 2,4,6-trinitrotoluene degradation: could we learn from (bio)chemistry for bioremediation and vice versa?Appl Microbiol Biotechnol. 2010 Nov;88(5):1043-64. doi: 10.1007/s00253-010-2830-x. Epub 2010 Sep 3. Appl Microbiol Biotechnol. 2010. PMID: 20814673 Review.
Cited by
-
Nicotinamide-independent asymmetric bioreduction of C=C-bonds via disproportionation of enones catalyzed by enoate reductases.Tetrahedron. 2010 Jan 16;66(3-2):663-667. doi: 10.1016/j.tet.2009.11.065. Tetrahedron. 2010. PMID: 21270958 Free PMC article.
-
A Rhizobium selenitireducens protein showing selenite reductase activity.Curr Microbiol. 2014 Mar;68(3):311-6. doi: 10.1007/s00284-013-0474-7. Epub 2013 Oct 25. Curr Microbiol. 2014. PMID: 24474405
-
Comparative characterization and expression analysis of the four Old Yellow Enzyme homologues from Shewanella oneidensis indicate differences in physiological function.Biochem J. 2006 Feb 15;394(Pt 1):335-44. doi: 10.1042/BJ20050979. Biochem J. 2006. PMID: 16293111 Free PMC article.
-
A Density Functional Theory Study toward Ring-Opening Reaction Mechanisms of 2,4,6-Trinitrotoluene's Meisenheimer Complex for the Biodegradation of Old Yellow Enzyme Flavoprotein Reductase.ACS Omega. 2020 Sep 4;5(37):23613-23620. doi: 10.1021/acsomega.0c02162. eCollection 2020 Sep 22. ACS Omega. 2020. PMID: 32984681 Free PMC article.
-
The engineer's approach to biology.EMBO Rep. 2006 Jan;7(1):21-3. doi: 10.1038/sj.embor.7400607. EMBO Rep. 2006. PMID: 16391532 Free PMC article.
References
-
- Achtnich, C., U. Sieglen, H.-J. Knackmuss, and H. Lenke. 1999. Irreversible binding of biologically reduced 2,4,6-trinitrotoluene to soil. Environ. Toxicol. Chem. 18:2416-2423.
-
- Ahmad, F., and J. B. Hughes. 2002. Reactivity of partially reduced arylhydroxylamine and nitrosoarene metabolites of 2,4,6-trinitrotoluene (TNT) toward biomass and humic acids. Environ. Sci. Technol. 36:4370-4381. - PubMed
-
- Barna, T., H. L. Messiha, C. Petosa, N. C. Bruce, N. S. Scrutton, and P. C. E. Moody. 2002. Crystal structure of bacterial morphinone reductase and properties of the C191A mutant enzyme. J. Biol. Chem. 277:30976-30983. - PubMed
-
- Barna, T. M., H. Khan, N. C. Bruce, I. Barsukov, N. S. Scrutton, and P. C. Moody. 2001. Crystal structure of pentaerythritol tetranitrate reductase: “flipped” binding geometries for steroid substrates in different redox states of the enzyme. J. Mol. Biol. 310:433-447. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

