Sequence-specific cleavage of small-subunit (SSU) rRNA with oligonucleotides and RNase H: a rapid and simple approach to SSU rRNA-based quantitative detection of microorganisms
- PMID: 15184170
- PMCID: PMC427779
- DOI: 10.1128/AEM.70.6.3650-3663.2004
Sequence-specific cleavage of small-subunit (SSU) rRNA with oligonucleotides and RNase H: a rapid and simple approach to SSU rRNA-based quantitative detection of microorganisms
Abstract
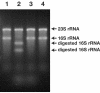
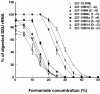
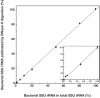
A rapid and simple approach to the small-subunit (SSU) rRNA-based quantitative detection of a specific group of microorganisms in complex ecosystems has been developed. The method employs sequence-specific cleavage of rRNA molecules with oligonucleotides and RNase H. Defined mixtures of SSU rRNAs were mixed with an oligonucleotide (referred to as a "scissor probe") that was specifically designed to hybridize with a particular site of targeted rRNA and were subsequently digested with RNase H to proceed to sequence-dependent rRNA scission at the hybridization site. Under appropriate reaction conditions, the targeted rRNAs were correctly cut into two fragments, whereas nontargeted rRNAs remained intact under the same conditions. The specificity of the cleavage could be properly adjusted by controlling the hybridization stringency between the rRNA and the oligonucleotides, i.e., by controlling either the temperature of the reaction or the formamide concentration in the hybridization-digestion buffer used for the reaction. This enabled the reliable discrimination of completely matched rRNA sequences from single-base mismatched sequences. For the detection of targeted rRNAs, the resulting RNA fragment patterns were analyzed by gel electrophoresis with nucleotide-staining fluorescent dyes in order to separate cleaved and intact rRNA molecules. The relative abundance of the targeted SSU rRNA fragments in the total SSU rRNA could easily be calculated without the use of an external standard by determining the signal intensity of individual SSU rRNA bands in the electropherogram. This approach provides a fast and easy means of identification, detection, and quantification of a particular group of microbes in clinical and environmental specimens based on rRNA.
Figures





References
-
- Afonina, E. I., N. V. Chichkova, and A. A. Bogdanov. 1991. RNA-RNA and RNA-protein interactions in 30S ribosomal subunits: association of 16S rRNA fragments in the presence of ribosomal proteins. FEBS Lett. 283:251-254. - PubMed
-
- Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridization. Curr. Opin. Biotechnol. 12:231-236. - PubMed
-
- Amann, R., and W. Ludwig. 2000. Ribosomal RNA-targeted nucleic acid probes for studies in microbial ecology. FEMS Microbiol. Rev. 24:555-565. - PubMed
-
- Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans and J. D. van Elass (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

