Regulation of a formin complex by the microtubule plus end protein tea1p
- PMID: 15184402
- PMCID: PMC2172381
- DOI: 10.1083/jcb.200403090
Regulation of a formin complex by the microtubule plus end protein tea1p
Abstract
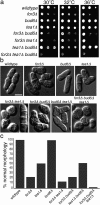
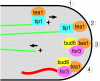
The plus ends of microtubules have been speculated to regulate the actin cytoskeleton for the proper positioning of sites of cell polarization and cytokinesis. In the fission yeast Schizosaccharomyces pombe, interphase microtubules and the kelch repeat protein tea1p regulate polarized cell growth. Here, we show that tea1p is directly deposited at cell tips by microtubule plus ends. Tea1p associates in large "polarisome" complexes with bud6p and for3p, a formin that assembles actin cables. Tea1p also interacts in a separate complex with the CLIP-170 protein tip1p, a microtubule plus end-binding protein that anchors tea1p to the microtubule plus end. Localization experiments suggest that tea1p and bud6p regulate formin distribution and actin cable assembly. Although single mutants still polarize, for3Deltabud6Deltatea1Delta triple-mutant cells lack polarity, indicating that these proteins contribute overlapping functions in cell polarization. Thus, these experiments begin to elucidate how microtubules contribute to the proper spatial regulation of actin assembly and polarized cell growth.
Copyright the Rockefeller University Press
Figures
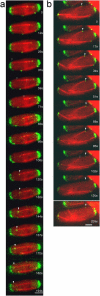
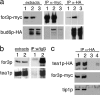

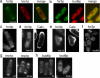
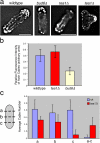
Similar articles
-
Regulation of actin assembly by microtubules in fission yeast cell polarity.Novartis Found Symp. 2005;269:59-66; discussion 66-72, 223-30. Novartis Found Symp. 2005. PMID: 16355535
-
Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast.Curr Biol. 2001 Jun 5;11(11):836-45. doi: 10.1016/s0960-9822(01)00235-4. Curr Biol. 2001. PMID: 11516644
-
Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity.Dev Cell. 2005 Apr;8(4):479-91. doi: 10.1016/j.devcel.2005.02.008. Dev Cell. 2005. PMID: 15809031
-
CLIP-170 family members: a motor-driven ride to microtubule plus ends.Dev Cell. 2004 Jun;6(6):746-8. doi: 10.1016/j.devcel.2004.05.017. Dev Cell. 2004. PMID: 15177023 Review.
-
Cell polarity: a tale of two Ts.Curr Biol. 2001 Aug 7;11(15):R600-2. doi: 10.1016/s0960-9822(01)00362-1. Curr Biol. 2001. PMID: 11516965 Review.
Cited by
-
Kelch repeat protein Clakel2p and calcium signaling control appressorium development in Colletotrichum lagenarium.Eukaryot Cell. 2008 Jan;7(1):102-11. doi: 10.1128/EC.00227-07. Epub 2007 Nov 26. Eukaryot Cell. 2008. PMID: 18039945 Free PMC article.
-
CLASP regulates mitochondrial distribution in Schizosaccharomyces pombe.J Cell Biol. 2008 Jul 14;182(1):41-9. doi: 10.1083/jcb.200712147. Epub 2008 Jul 7. J Cell Biol. 2008. PMID: 18606849 Free PMC article.
-
Redundant mechanisms recruit actin into the contractile ring in silkworm spermatocytes.PLoS Biol. 2008 Sep 2;6(9):e209. doi: 10.1371/journal.pbio.0060209. PLoS Biol. 2008. PMID: 18767903 Free PMC article.
-
Structure of the formin-interaction domain of the actin nucleation-promoting factor Bud6.Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3424-33. doi: 10.1073/pnas.1203035109. Epub 2012 Nov 16. Proc Natl Acad Sci U S A. 2012. PMID: 23161908 Free PMC article.
-
Establishing new sites of polarization by microtubules.Curr Biol. 2009 Jan 27;19(2):83-94. doi: 10.1016/j.cub.2008.12.008. Epub 2009 Jan 15. Curr Biol. 2009. PMID: 19147354 Free PMC article.
References
-
- Adams, J., R. Kelso, and L. Cooley. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10:17–24. - PubMed
-
- Browning, H., D.D. Hackney, and P. Nurse. 2003. Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5:812–818. - PubMed
-
- Brunner, D., and P. Nurse. 2000. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 102:695–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

