Characterization of Chlamydia trachomatis omp1 genotypes detected in eye swab samples from remote Australian communities
- PMID: 15184427
- PMCID: PMC427869
- DOI: 10.1128/JCM.42.6.2501-2507.2004
Characterization of Chlamydia trachomatis omp1 genotypes detected in eye swab samples from remote Australian communities
Abstract
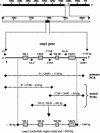
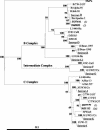
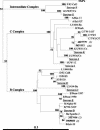
Chlamydia trachomatis conjunctival samples collected over a 6-month period from individuals with clinical signs of trachoma and located in remote communities in the Australian Northern Territory were differentially characterized according to serovar and variants. The rationale was to gain an understanding of the epidemiology of an apparent increased prevalence of acute trachoma in areas thought to be less conducive to this disease. Characterization was performed through sequencing of a region of the omp1 gene spanning the four variable domains and encoding the major outer membrane protein. Nucleotide and deduced amino acid sequences were genotyped by using a BLAST similarity search and were examined by phylogenetic analyses to illustrate evolutionary relationships between the clinical and GenBank reference strains. The predominant genotype identified corresponded to that of serovar C (87.1%), followed by the genotype corresponding to serovar Ba (12.9%). All nucleotide and amino acid sequences exhibited minor levels of variation with respect to GenBank reference sequences. The omp1 nucleotide sequences of the clinical samples best aligned with those of the conjunctival C. trachomatis reference strains C/TW-3/OT and Ba/Apache-2. All clinical samples (of serovar C) exhibited four or five nucleotide changes compared with C/TW-3/OT, while all serovar Ba samples had one or two nucleotide differences from Ba/Apache-2. Phylogenetic analyses revealed close relationships between these Northern Territory chlamydial samples and the respective reference strains, although the high proportion of sequence variants suggests an evolutionarily distinct C. trachomatis population causing eye infections in Australia. Given that such genotypic information has gone unreported, these findings provide knowledge and a foundation for trachoma-associated C. trachomatis variants circulating in the Northern Territory.
Figures
References
-
- Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. - PMC - PubMed
-
- Bowden, F. J., B. A. Paterson, J. Mein, J. Savage, C. K. Fairley, S. M. Garland, and S. N. Tabrizi. 1999. Estimating the prevalence of Trichomonas vaginalis, Chlamydia trachomatis, Neisseria gonorrhoeae, and human papillomavirus infection in indigenous women in northern Australia. Sex. Transm. Infect. 75:431-434. - PMC - PubMed
-
- Cabral, T., A. M. Jolly, and J. L. Wylie. 2003. Chlamydia trachomatis omp1 genotypic diversity and concordance with sexual network data. J. Infect. Dis. 187:279-286. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

