Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types
- PMID: 15184441
- PMCID: PMC427854
- DOI: 10.1128/JCM.42.6.2609-2617.2004
Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types
Abstract
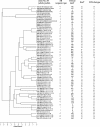
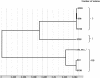
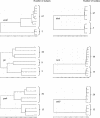
A multilocus sequence typing (MLST) scheme was developed to study the genetic relationships and population structure of 72 Clostridium difficile isolates from various hosts, geographic sources, PCR ribotypes, and toxigenic types (determined by PCR targeting tcdA and tcdB genes). MLST was performed by DNA sequence analysis of seven housekeeping genes (aroE, ddl, dutA, tpi, recA, gmk, and sodA). The number of alleles ranged from five (dutA and ddl) to eleven (recA). Allelic profiles allowed the definition of 34 different sequence types (STs). These STs lacked correlation with geographic source but were well correlated to toxigenic type. The dendrogram generated from a matrix of pairwise genetic distances showed that animal isolates did not constitute a distinct lineage from human isolates and that there was no hypervirulent lineage within the population of toxigenic human isolates (isolates recovered from pseudomembranous colitis and antibiotic-associated diarrhea did not cluster in distinct lineages). However, A(-) B(+) variant isolates shared the same ST that appeared as a divergent lineage in the population studied, indicating a single evolutionary origin. The population structure was further examined by analysis of allelic polymorphism. The dendrogram generated from composite sequence-based analysis revealed a homogeneous population associated with three divergent lineages, one of which was restricted to A(-) B(+) variant isolates. C. difficile exhibited a clonal population structure, as revealed by the estimation of linkage disequilibrium (Ia) between loci. The analysis of alleles within clonal complexes estimated that point mutation generated new alleles at a frequency eightfold higher than recombinational exchange, and the congruence of the dendrograms generated from separate housekeeping loci confirmed the mutational evolution of this species.
Figures
References
-
- Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. - PMC - PubMed
-
- Barbut, F., N. Mario, M. Delmee, J. Gozian, and J. C. Petit. 1993. Genomic fingerprinting of Clostridium difficile isolates by using a random amplified polymorphic DNA (RAPD) assay. FEMS Microbiol. Lett. 114:161-166. - PubMed
-
- Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778-782. - PubMed
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

