Identification of subtype C human immunodeficiency virus type 1 by subtype-specific PCR and its use in the characterization of viruses circulating in the southern parts of India
- PMID: 15184461
- PMCID: PMC427845
- DOI: 10.1128/JCM.42.6.2742-2751.2004
Identification of subtype C human immunodeficiency virus type 1 by subtype-specific PCR and its use in the characterization of viruses circulating in the southern parts of India
Abstract
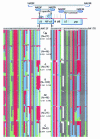
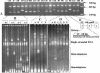
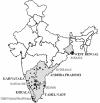
Human immunodeficiency virus type 1 (HIV-1) subtype C viruses are associated with nearly half of worldwide HIV-1 infections and are most predominant in India and the southern and eastern parts of Africa. Earlier reports from India identified the preponderance of subtype C and a small proportion of subtype A viruses. Subsequent reports identifying multiple subtypes suggest new introductions and/or their detection due to extended screening. The southern parts of India constitute emerging areas of the epidemic, but it is not known whether HIV-1 infection in these areas is associated with subtype C viruses or is due to the potential new introduction of non-subtype C viruses. Here, we describe the development of a specific and sensitive PCR-based strategy to identify subtype C-viruses (C-PCR). The strategy is based on amplifying a region encompassing a long terminal repeat and gag in the first round, followed by two sets of nested primers; one amplifies multiple subtypes, while the other is specific to subtype C. The common HIV and subtype C-specific fragments are distinguishable by length differences in agarose gels and by the difference in the numbers of NF-kappaB sites encoded in the subtype C-specific fragment. We implemented this method to screen 256 HIV-1-infected individuals from 35 towns and cities in four states in the south and a city in the east. With the exception of single samples of subtypes A and B and a B/C recombinant, we found all to be infected with subtype C viruses, and the subtype assignments were confirmed in a subset by using heteroduplex mobility assays and phylogenetic analysis of sequences. We propose the use of C-PCR to facilitate rapid molecular epidemiologic characterization to aid vaccine and therapeutic strategies.
Figures


Similar articles
-
[Development of a subtype screening assay for human immunodeficiency virus type 1 by nested multiplex PCR].Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2004 Mar;18(1):83-7. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2004. PMID: 15340536 Chinese.
-
Restriction fragment length polymorphism analysis for rapid gag subtype determination of human immunodeficiency virus type 1 in South Africa.J Virol Methods. 1999 Mar;78(1-2):51-9. doi: 10.1016/s0166-0934(98)00163-3. J Virol Methods. 1999. PMID: 10204696
-
Characterization of a new circulating recombinant form comprising HIV-1 subtypes C and B in southern Brazil.AIDS. 2006 Oct 24;20(16):2011-9. doi: 10.1097/01.aids.0000247573.95880.db. AIDS. 2006. PMID: 17053347
-
Simple subtyping assay for human immunodeficiency virus type 1 subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC.J Clin Microbiol. 2004 Sep;42(9):4261-7. doi: 10.1128/JCM.42.9.4261-4267.2004. J Clin Microbiol. 2004. PMID: 15365021 Free PMC article.
-
Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents.AIDS Res Hum Retroviruses. 2001 Jan 20;17(2):161-8. doi: 10.1089/08892220150217247. AIDS Res Hum Retroviruses. 2001. PMID: 11177395
Cited by
-
Role of HIV-1 subtype C envelope V3 to V5 regions in viral entry, coreceptor utilization and replication efficiency in primary T-lymphocytes and monocyte-derived macrophages.Virol J. 2007 Nov 24;4:126. doi: 10.1186/1743-422X-4-126. Virol J. 2007. PMID: 18036244 Free PMC article.
-
Differential induction of rat neuronal excitotoxic cell death by human immunodeficiency virus type 1 clade B and C tat proteins.AIDS Res Hum Retroviruses. 2011 Jun;27(6):647-54. doi: 10.1089/AID.2010.0192. Epub 2010 Dec 7. AIDS Res Hum Retroviruses. 2011. PMID: 20977378 Free PMC article.
-
HLA-B57 and gender influence the occurrence of tuberculosis in HIV infected people of south India.Clin Dev Immunol. 2011;2011:549023. doi: 10.1155/2011/549023. Epub 2011 Aug 25. Clin Dev Immunol. 2011. PMID: 21876708 Free PMC article.
-
Flail arm-like syndrome associated with HIV-1 infection.Ann Indian Acad Neurol. 2009 Apr;12(2):127-30. doi: 10.4103/0972-2327.53084. Ann Indian Acad Neurol. 2009. PMID: 20142861 Free PMC article.
-
Near full-length genomic characterization of a HIV type 1 BC recombinant strain from Manipur, India.Virus Genes. 2012 Oct;45(2):201-6. doi: 10.1007/s11262-012-0768-z. Epub 2012 Jun 19. Virus Genes. 2012. PMID: 22710995
References
-
- Bongertz, V., D. C. Bou-Habib, L. F. Brigido, M. Caseiro, P. J. Chequer, J. C. Couto-Fernandez, P. C. Ferreira, B. Galvao-Castro, D. Greco, M. L. Guimaraes, M. I. Linhares de Carvalho, M. G. Morgado, C. A. Oliveira, S. Osmanov, C. A. Ramos, M. Rossini, E. Sabino, A. Tanuri, M. Ueda, et al. 2000. HIV-1 diversity in Brazil: genetic, biologic, and immunologic characterization of HIV-1 strains in three potential HIV vaccine evaluation sites. J. Acquir. Immun. Defic. Syndr. 23:184-193. - PubMed
-
- Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, M. O'Shaughnessy, and C. Y. Ou. 1996. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. - PubMed
-
- Cham, F., L. Heyndrickx, W. Janssens, K. Vereecken, K. De Houwer, S. Coppens, G. Van der Auwera, H. Whittle, and G. van Der Groen. 2000. Development of a one-tube multiplex reverse transcriptase-polymerase chain reaction assay for the simultaneous amplification of HIV type 1 group M gag and env heteroduplex mobility assay fragments. AIDS Res. Hum. Retrovir. 16:1503-1505. - PubMed
-
- Choudhury, S., M. A. Montano, C. Womack, J. T. Blackard, J. K. Maniar, D. G. Saple, S. Tripathy, S. Sahni, S. Shah, G. P. Babu, and M. Essex. 2000. Increased promoter diversity reveals a complex phylogeny of human immunodeficiency virus type 1 subtype C in India. J. Hum. Virol. 3:35-43. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

