Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA
- PMID: 15184654
- PMCID: PMC428464
- DOI: 10.1073/pnas.0403090101
Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA
Abstract
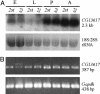
Adaptive changes in nature occur by a variety of mechanisms, and Drosophila chromosomal inversions was one of the first studied examples. However, the precise genetic causes of the adaptive value of inversions remain uncertain. Here we investigate the impact of the widespread inversion 2j of Drosophila buzzatii on the expression of the CG13617 gene, whose coding region is located only 12 bp away from the inversion proximal breakpoint. This gene is transcribed into a 2.3-kb mRNA present in all D. buzzatii developmental stages. More importantly, the expression level of CG13617 is reduced 5-fold in embryos of lines homozygous for the 2j inversion compared with lines without the inversion. An antisense RNA that originates in the Foldback-like transposon Kepler inserted at the breakpoint junction in all of the 2j lines and that forms duplexes with the CG13617 mRNA in 2j embryos is most likely responsible for the near silencing of the gene. Few examples of RNA interference caused by transposable elements (TEs) have been previously described, but this mechanism might be prevalent in many organisms and illustrates the potential of TEs as a major source of genetic variation. In addition, because chromosomal rearrangements are usually induced by TEs, position effects might be more common than previously recognized and contribute significantly to the evolutionary success of inversions.
Figures



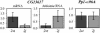

References
-
- Krimbas, C. B. & Powell, J. R., eds. (1992) in Drosophila Inversion Polymorphism (CRC, Boca Raton, FL), pp. 1-52.
-
- Lewontin, R. C., Moore, J. A., Provine, W. B. & Wallace, B. (1981) Dobzhansky's Genetics of Natural Populations, I-XLIII (Columbia Univ. Press, New York).
-
- Dobzhansky, T. (1970) Genetics of the Evolutionary Process (Columbia Univ. Press, New York).
-
- Charlesworth, B. (1974) Genet. Res. 23, 259-280. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

