Signal sequence analysis of expressed sequence tags from the nematode Nippostrongylus brasiliensis and the evolution of secreted proteins in parasites
- PMID: 15186490
- PMCID: PMC463072
- DOI: 10.1186/gb-2004-5-6-r39
Signal sequence analysis of expressed sequence tags from the nematode Nippostrongylus brasiliensis and the evolution of secreted proteins in parasites
Abstract
Background: Parasitism is a highly successful mode of life and one that requires suites of gene adaptations to permit survival within a potentially hostile host. Among such adaptations is the secretion of proteins capable of modifying or manipulating the host environment. Nippostrongylus brasiliensis is a well-studied model nematode parasite of rodents, which secretes products known to modulate host immunity.
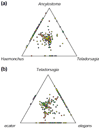
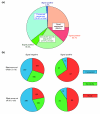
Results: Taking a genomic approach to characterize potential secreted products, we analyzed expressed sequence tag (EST) sequences for putative amino-terminal secretory signals. We sequenced ESTs from a cDNA library constructed by oligo-capping to select full-length cDNAs, as well as from conventional cDNA libraries. SignalP analysis was applied to predicted open reading frames, to identify potential signal peptides and anchors. Among 1,234 ESTs, 197 (~16%) contain predicted 5' signal sequences, with 176 classified as conventional signal peptides and 21 as signal anchors. ESTs cluster into 742 distinct genes, of which 135 (18%) bear predicted signal-sequence coding regions. Comparisons of clusters with homologs from Caenorhabditis elegans and more distantly related organisms reveal that the majority (65% at P < e-10) of signal peptide-bearing sequences from N. brasiliensis show no similarity to previously reported genes, and less than 10% align to conserved genes recorded outside the phylum Nematoda. Of all novel sequences identified, 32% contained predicted signal peptides, whereas this was the case for only 3.4% of conserved genes with sequence homologies beyond the Nematoda.
Conclusions: These results indicate that secreted proteins may be undergoing accelerated evolution, either because of relaxed functional constraints, or in response to stronger selective pressure from host immunity.
Figures
References
-
- Lightowlers MW, Rickard MD. Excretory-secretory products of helminth parasites: effects on host immune responses. Parasitology. 1988;96:S123–S166. - PubMed
-
- Yatsuda AP, Krijgsveld J, Cornelissen AWCA, Heck AJ, De Vries E. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J Biol Chem. 2003;278:16941–16951. doi: 10.1074/jbc.M212453200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

