A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults
- PMID: 15186491
- PMCID: PMC463073
- DOI: 10.1186/gb-2004-5-6-r40
A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults
Abstract
Background: Sexual dimorphism results in the formation of two types of individuals with specialized reproductive roles and is most evident in the germ cells and gonads.
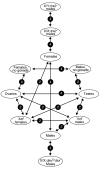
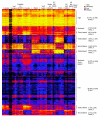
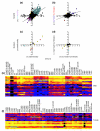
Results: We have undertaken a global analysis of transcription between the sexes using a 31,464 element FlyGEM microarray to determine what fraction of the genome shows sex-biased expression, what tissues express these genes, the predicted functions of these genes, and where these genes map onto the genome. Females and males (both with and without gonads), dissected testis and ovary, females and males with genetically ablated germlines, and sex-transformed flies were sampled.
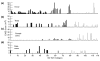
Conclusions: Using any of a number of criteria, we find extensive sex-biased expression in adults. The majority of cases of sex differential gene expression are attributable to the germ cells. There is also a large class of genes with soma-biased expression. There is little germline-biased expression indicating that nearly all genes with germline expression also show sex-bias. Monte Carlo simulations show that some genes with sex-biased expression are non-randomly distributed in the genome.
Figures




References
-
- McKeown M, Belote JM, Boggs RT. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988;53:887–895. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

