Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis
- PMID: 15186493
- PMCID: PMC463075
- DOI: 10.1186/gb-2004-5-6-r42
Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis
Abstract
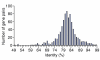
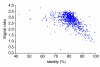
By comparing two fully sequenced genomes of Chlamydia trachomatis using competitive hybridization on DNA microarrays, a logarithmic correlation was demonstrated between the signal ratio of the arrays and the 75-99% range of nucleotide identities of the genes. Variable genes within 14 uncharacterized strains of C. trachomatis were identified by array analysis and verified by DNA sequencing. These genes may be crucial for understanding chlamydial virulence and pathogenesis.
Figures
References
-
- Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, Barrell BG, Parkhill J, Stoker NG, Karlyshev AV, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

