Transactivating effect of hepatitis C virus core protein: a suppression subtractive hybridization study
- PMID: 15188498
- PMCID: PMC4572261
- DOI: 10.3748/wjg.v10.i12.1746
Transactivating effect of hepatitis C virus core protein: a suppression subtractive hybridization study
Abstract
Aim: To investigate the transactivating effect of hepatitis C virus (HCV) core protein and to screen genes transactivated by HCV core protein.
Methods: pcDNA3.1(-)-core containing full-length HCV core gene was constructed by insertion of HCV core gene into EcoRI/BamHI site. HepG2 cells were cotransfected with pcDNA3.1(-)-core and pSV-lacZ. After 48 h, cells were collected and detected for the expression of beta-gal by an enzyme-linked immunosorbent assay (ELISA) kit. HepG2 cells were transiently transfected with pcDNA3.1(-)-core using Lipofectamine reagent. Cells were collected and total mRNA was isolated. A subtracted cDNA library was generated and constructed into a pGEM-Teasy vector. The library was amplified with E. coli strain JM109. The cDNAs were sequenced and analyzed in GenBank with BLAST search after polymerase chain reaction (PCR).
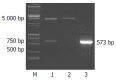
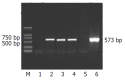
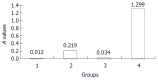
Results: The core mRNA and protein could be detected in HepG2 cell lysate which was transfected by the pcDNA3.1(-)-core. The activity of beta-galactosidase in HepG2 cells transfected by the pcDNA3.1(-)-core was 5.4 times higher than that of HepG2 cells transfected by control plasmid. The subtractive library of genes transactivated by HCV core protein was constructed successfully. The amplified library contained 233 positive clones. Colony PCR showed that 213 clones contained 100-1 000 bp inserts. Sequence analysis was performed in 63 clones. Six of the sequences were unknown genes. The full length sequences were obtained with bioinformatics method, accepted by GenBank. It was suggested that six novel cDNA sequences might be target genes transactivated by HCV core protein.
Conclusion: The core protein of HCV has transactivating effects on SV40 early promoter/enhancer. A total of 63 clones from cDNA library were randomly chosen and sequenced. Using the BLAST program at the National Center for Biotechnology Information, six of the sequences were unknown genes. The other 57 sequences were highly similar to known genes.
Figures

Similar articles
-
Transactivating effect of complete S protein of hepatitis B virus and cloning of genes transactivated by complete S protein using suppression subtractive hybridization technique.World J Gastroenterol. 2005 Jul 7;11(25):3893-8. doi: 10.3748/wjg.v11.i25.3893. World J Gastroenterol. 2005. PMID: 15991289 Free PMC article.
-
Study of transactivating effect of pre-S2 protein of hepatitis B virus and cloning of genes transactivated by pre-S2 protein with suppression subtractive hybridization.World J Gastroenterol. 2005 Sep 21;11(35):5438-43. doi: 10.3748/wjg.v11.i35.5438. World J Gastroenterol. 2005. PMID: 16222733 Free PMC article.
-
[Screening and cloning target genes transactivated by hepatitis C virus F protein using suppression subtractive hybridization technique].Zhonghua Gan Zang Bing Za Zhi. 2005 Sep;13(9):660-3. Zhonghua Gan Zang Bing Za Zhi. 2005. PMID: 16174453 Chinese.
-
Screening and cloning for proteins transactivated by the PS1TP5 protein of hepatitis B virus: a suppression subtractive hybridization study.World J Gastroenterol. 2007 Mar 14;13(10):1602-7. doi: 10.3748/wjg.v13.i10.1602. World J Gastroenterol. 2007. PMID: 17461456 Free PMC article.
-
[Cloning of genes transactivated by nonstructural protein 4A of hepatitis C virus.].Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2006 Sep;20(3):270-2. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2006. PMID: 17086291 Chinese.
Cited by
-
Screening of hepatocyte proteins binding to F protein of hepatitis C virus by yeast two-hybrid system.World J Gastroenterol. 2005 Sep 28;11(36):5659-65. doi: 10.3748/wjg.v11.i36.5659. World J Gastroenterol. 2005. PMID: 16237761 Free PMC article.
-
Cyclic adenosine monophosphate response element-binding protein transcriptionally regulates CHCHD2 associated with the molecular pathogenesis of hepatocellular carcinoma.Mol Med Rep. 2015 Jun;11(6):4053-62. doi: 10.3892/mmr.2015.3256. Epub 2015 Jan 26. Mol Med Rep. 2015. PMID: 25625293 Free PMC article.
-
A chemical proteomics approach for global analysis of lysine monomethylome profiling.Mol Cell Proteomics. 2015 Feb;14(2):329-39. doi: 10.1074/mcp.M114.044255. Epub 2014 Dec 11. Mol Cell Proteomics. 2015. PMID: 25505155 Free PMC article.
References
-
- Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362] J Hepatol. 2002;36:582–585. - PubMed
-
- Di Bisceglie AM. Hepatitis C and hepatocellular carcinoma. Hepatology. 1997;26(3 Suppl 1):34S–38S. - PubMed
-
- Hasan F, Jeffers LJ, De Medina M, Reddy KR, Parker T, Schiff ER, Houghton M, Choo QL, Kuo G. Hepatitis C-associated hepatocellular carcinoma. Hepatology. 1990;12(3 Pt 1):589–591. - PubMed
-
- Ito T, Mukaigawa J, Zuo J, Hirabayashi Y, Mitamura K, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77(Pt 5):1043–1054. - PubMed
-
- Clarke B. Molecular virology of hepatitis C virus. J Gen Virol. 1997;78(Pt 10):2397–2410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

