Prediction of charge-induced molecular alignment of biomolecules dissolved in dilute liquid-crystalline phases
- PMID: 15189846
- PMCID: PMC1304251
- DOI: 10.1529/biophysj.103.035790
Prediction of charge-induced molecular alignment of biomolecules dissolved in dilute liquid-crystalline phases
Abstract
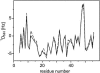
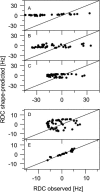
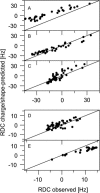
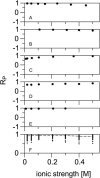
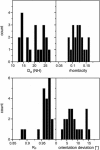
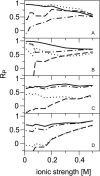
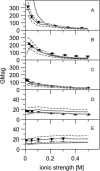
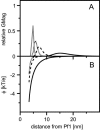
Alignment of macromolecules in nearly neutral aqueous lyotropic liquid-crystalline media such as bicelles, commonly used in macromolecular NMR studies, can be predicted accurately by a steric obstruction model (Zweckstetter and Bax, 2000). A simple extension of this model is described that results in improved predictions for both the alignment orientation and magnitude of protein and DNA solutes in charged nematic media, such as the widely used medium of filamentous phage Pf1. The extended model approximates the electrostatic interaction between a solute and an ordered phage particle as that between the solute's surface charges and the electric field of the phage. The model is evaluated for four different proteins and a DNA oligomer. Results indicate that alignment in charged nematic media is a function not only of the solute's shape, but also of its electric multipole moments of net charge, dipole, and quadrupole. The relative importance of these terms varies greatly from one macromolecule to another, and evaluation of the experimental data indicates that these terms scale differently with ionic strength. For several of the proteins, the calculated alignment is sensitive to the precise position of the charged groups on the protein surface. This suggests that NMR alignment measurements can potentially be used to probe protein electrostatics. Inclusion of electrostatic interactions in addition to steric effects makes the extended model applicable to all liquid crystals used in biological NMR to date.
Figures

References
-
- Al-Hashimi, H. M., H. Valafar, M. Terrell, E. R. Zartler, M. K. Eidsness, and J. H. Prestegard. 2000. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J. Magn. Reson. 143:402–406. - PubMed
-
- Almond, A., and J. B. Axelsen. 2002. Physical interpretation of residual dipolar couplings in neutral aligned media. J. Am. Chem. Soc. 124:9986–9987. - PubMed
-
- Antosiewicz, J., J. A. McCammon, and M. K. Gilson. 1994. Prediction of pH-dependent properties of proteins. J. Mol. Biol. 238:415–436. - PubMed
-
- Azurmendi, H. F., and C. A. Bush. 2002. Conformational studies of blood group A and blood group B oligosaccharides using NMR residual dipolar couplings. Carbohydr. Res. 337:905–915. - PubMed
-
- Barrientos, L. G., C. Dolan, and A. M. Gronenborn. 2000. Characterization of surfactant liquid crystal phases suitable for molecular alignment and measurement of dipolar couplings. J. Biomol. NMR. 16:329–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

