The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae
- PMID: 15189985
- PMCID: PMC420139
- DOI: 10.1128/EC.3.3.632-645.2004
The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae
Abstract
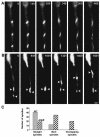
Kinesins are motor proteins which are classified into 11 different families. We identified 11 kinesin-like proteins in the genome of the filamentous fungus Aspergillus nidulans. Relatedness analyses based on the motor domains grouped them into nine families. In this paper, we characterize KipB as a member of the Kip3 family of microtubule depolymerases. The closest homologues of KipB are Saccharomyces cerevisiae Kip3 and Schizosaccharomyces pombe Klp5 and Klp6, but sequence similarities outside the motor domain are very low. A disruption of kipB demonstrated that it is not essential for vegetative growth. kipB mutant strains were resistant to high concentrations of the microtubule-destabilizing drug benomyl, suggesting that KipB destabilizes microtubules. kipB mutations caused a failure of spindle positioning in the cell, a delay in mitotic progression, an increased number of bent mitotic spindles, and a decrease in the depolymerization of cytoplasmic microtubules during interphase and mitosis. Meiosis and ascospore formation were not affected. Disruption of the kipB gene was synthetically lethal in combination with the temperature-sensitive mitotic kinesin motor mutation bimC4, suggesting an important but redundant role of KipB in mitosis. KipB localized to cytoplasmic, astral, and mitotic microtubules in a discontinuous pattern, and spots of green fluorescent protein moved along microtubules toward the plus ends.
Copyright 2004 American Society for Microbiology
Figures







Similar articles
-
Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast.Mol Biol Cell. 2001 Dec;12(12):3919-32. doi: 10.1091/mbc.12.12.3919. Mol Biol Cell. 2001. PMID: 11739790 Free PMC article.
-
Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics.Mol Biol Cell. 2008 Dec;19(12):5104-15. doi: 10.1091/mbc.e08-02-0224. Epub 2008 Sep 17. Mol Biol Cell. 2008. PMID: 18799626 Free PMC article.
-
Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans.J Cell Sci. 2005 Aug 15;118(Pt 16):3705-16. doi: 10.1242/jcs.02501. J Cell Sci. 2005. PMID: 16105883
-
Mitotic motors in Saccharomyces cerevisiae.Biochim Biophys Acta. 2000 Mar 17;1496(1):99-116. doi: 10.1016/s0167-4889(00)00012-4. Biochim Biophys Acta. 2000. PMID: 10722880 Review.
-
CLIP-170 family members: a motor-driven ride to microtubule plus ends.Dev Cell. 2004 Jun;6(6):746-8. doi: 10.1016/j.devcel.2004.05.017. Dev Cell. 2004. PMID: 15177023 Review.
Cited by
-
Kinesin Motors in the Filamentous Basidiomycetes in Light of the Schizophyllum commune Genome.J Fungi (Basel). 2022 Mar 12;8(3):294. doi: 10.3390/jof8030294. J Fungi (Basel). 2022. PMID: 35330296 Free PMC article. Review.
-
A Tubulin Binding Switch Underlies Kip3/Kinesin-8 Depolymerase Activity.Dev Cell. 2017 Jul 10;42(1):37-51.e8. doi: 10.1016/j.devcel.2017.06.011. Dev Cell. 2017. PMID: 28697331 Free PMC article.
-
Move in for the kill: motile microtubule regulators.Trends Cell Biol. 2012 Nov;22(11):567-75. doi: 10.1016/j.tcb.2012.08.003. Epub 2012 Sep 6. Trends Cell Biol. 2012. PMID: 22959403 Free PMC article. Review.
-
Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement.Mol Biol Cell. 2009 Feb;20(3):963-72. doi: 10.1091/mbc.e08-09-0979. Epub 2008 Nov 26. Mol Biol Cell. 2009. PMID: 19037096 Free PMC article.
-
Three-Dimensional Optical Tweezers Tracking Resolves Random Sideward Steps of the Kinesin-8 Kip3.Biophys J. 2018 Nov 20;115(10):1993-2002. doi: 10.1016/j.bpj.2018.09.026. Epub 2018 Oct 2. Biophys J. 2018. PMID: 30360926 Free PMC article.
References
-
- Aist, J. R., and N. R. Morris. 1999. Mitosis in filamentous fungi: how we got where we are. Fungal Genet. Biol. 27:1-25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

