Inducible defense mechanism against nitric oxide in Candida albicans
- PMID: 15189992
- PMCID: PMC420131
- DOI: 10.1128/EC.3.3.715-723.2004
Inducible defense mechanism against nitric oxide in Candida albicans
Abstract
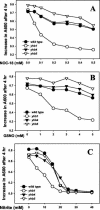
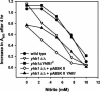
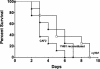
The yeast Candida albicans is an opportunistic pathogen that threatens patients with compromised immune systems. Immune cell defenses against C. albicans are complex but typically involve the production of reactive oxygen species and nitrogen radicals such as nitric oxide (NO) that damage the yeast or inhibit its growth. Whether Candida defends itself against NO and the molecules responsible for this defense have yet to be determined. The defense against NO in various bacteria and the yeast Saccharomyces cerevisiae involves an NO-scavenging flavohemoglobin. The C. albicans genome contains three genes encoding flavohemoglobin-related proteins, CaYHB1, CaYHB4, and CaYHB5. To assess their roles in NO metabolism, we constructed strains lacking each of these genes and demonstrated that just one, CaYHB1, is responsible for NO consumption and detoxification. In C. albicans, NO metabolic activity and CaYHB1 mRNA levels are rapidly induced by NO and NO-generating agents. Loss of CaYHB1 increases the sensitivity of C. albicans to NO-mediated growth inhibition. In mice, infections with Candida strains lacking CaYHB1 still resulted in lethality, but virulence was decreased compared to that in wild-type strains. Thus, C. albicans possesses a rapid, specific, and highly inducible NO defense mechanism involving one of three putative flavohemoglobin genes.
Copyright 2004 American Society for Microbiology
Figures



References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. Green Publishing Associates and Wiley-Interscience, New York, N.Y.
-
- Bain, J. M., C. Stubberfield, and N. A. Gow. 2001. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol. Lett. 204:323-328. - PubMed
-
- Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

