Sequence elements necessary for transcriptional activation of BAD1 in the yeast phase of Blastomyces dermatitidis
- PMID: 15189999
- PMCID: PMC420126
- DOI: 10.1128/EC.3.3.785-794.2004
Sequence elements necessary for transcriptional activation of BAD1 in the yeast phase of Blastomyces dermatitidis
Abstract
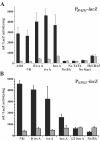
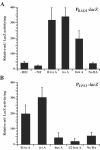
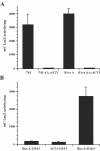
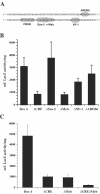
Blastomyces dermatitidis is a dimorphic fungal pathogen that converts from mycelia or conidia to a host-adapted yeast morphotype upon infection. Conversion to the yeast form is accompanied by the production of the virulence factor BAD1. Yeast-phase-specific expression of BAD1 is transcriptionally regulated, and its promoter shares homology with that of the yeast-phase-specific gene YPS3 of Histoplasma capsulatum. Serial truncations of the BAD1 upstream region were fused to the lacZ reporter to define functional areas in the promoter. Examination of PBAD1-lacZ fusions in B. dermatitidis indicated that BAD1 transcription is upregulated in the yeast phase. The 63-nucleotide box A region conserved in the YPS3 upstream region was shown to be an essential component of the minimal BAD1 promoter. A matched PYPS3-lacZ construct indicated that this same region was needed for minimal YPS3 promoter activity in B. dermatitidis transformants. Reporter activity in H. capsulatum transformants similarly showed a requirement for box A in the minimal BAD1 promoter. Several putative transcription factor binding sites were identified within box A of BAD1. Replacement of two of these predicted sites within box A--a cAMP responsive element and a Myb binding site--sharply reduced transcriptional activity, indicating that these regions are critical in dictating the yeast-phase-specific expression of this crucial virulence determinant of B. dermatitidis.
Copyright 2004 American Society for Microbiology
Figures

Similar articles
-
Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism.Mol Microbiol. 2001 Feb;39(4):875-89. doi: 10.1046/j.1365-2958.2001.02300.x. Mol Microbiol. 2001. PMID: 11251809
-
Molecular genetic analysis of Blastomyces dermatitidis reveals new insights about pathogenic mechanisms.Int J Med Microbiol. 2002 Oct;292(5-6):363-71. doi: 10.1078/1438-4221-00219. Int J Med Microbiol. 2002. PMID: 12452282 Review.
-
A C-terminal EGF-like domain governs BAD1 localization to the yeast surface and fungal adherence to phagocytes, but is dispensable in immune modulation and pathogenicity of Blastomyces dermatitidis.Mol Microbiol. 2003 Apr;48(1):53-65. doi: 10.1046/j.1365-2958.2003.03415.x. Mol Microbiol. 2003. PMID: 12657044
-
Role of glucan and surface protein BAD1 in complement activation by Blastomyces dermatitidis yeast.Infect Immun. 2001 Dec;69(12):7559-64. doi: 10.1128/IAI.69.12.7559-7564.2001. Infect Immun. 2001. PMID: 11705933 Free PMC article.
-
Using new genetic tools to study the pathogenesis of Blastomyces dermatitidis.Trends Microbiol. 2002 Jan;10(1):25-30. doi: 10.1016/s0966-842x(01)02258-2. Trends Microbiol. 2002. PMID: 11755082 Review.
Cited by
-
Revisiting old friends: Developments in understanding Histoplasma capsulatum pathogenesis.J Microbiol. 2016 Mar;54(3):265-76. doi: 10.1007/s12275-016-6044-5. Epub 2016 Feb 27. J Microbiol. 2016. PMID: 26920886 Review.
-
Histoplasma yeast and mycelial transcriptomes reveal pathogenic-phase and lineage-specific gene expression profiles.BMC Genomics. 2013 Oct 10;14:695. doi: 10.1186/1471-2164-14-695. BMC Genomics. 2013. PMID: 24112604 Free PMC article.
-
Surface localization of the Yps3p protein of Histoplasma capsulatum.Eukaryot Cell. 2005 Apr;4(4):685-93. doi: 10.1128/EC.4.4.685-693.2005. Eukaryot Cell. 2005. PMID: 15821128 Free PMC article.
-
Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis.PLoS Genet. 2011 Oct;7(10):e1002345. doi: 10.1371/journal.pgen.1002345. Epub 2011 Oct 27. PLoS Genet. 2011. PMID: 22046142 Free PMC article.
-
Cbp1, a fungal virulence factor under positive selection, forms an effector complex that drives macrophage lysis.PLoS Pathog. 2022 Jun 22;18(6):e1010417. doi: 10.1371/journal.ppat.1010417. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35731824 Free PMC article.
References
-
- Beijersbergen, A., A. den Dulk-Ras, R. A. Schilperoort, and P. J. J. Hooykaas. 1992. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324-1327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

