Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI
- PMID: 15190132
- PMCID: PMC434427
- DOI: 10.1093/nar/gkh618
Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI
Abstract
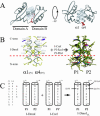
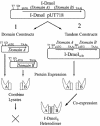
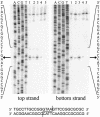
The general structural fold of the LAGLIDADG endonuclease family consists of two similar alpha/beta domains (alphabetabetaalphabetabetaalpha) that assemble either as homodimers or monomers with the domains related by pseudo-two-fold symmetry. At the center of this symmetry is the closely packed LAGLIDADG two-helix bundle that forms the main inter- or intra-molecular contact region between the domains of single- or double-motif proteins, respectively. In this work, we further examine the role of the LAGLIDADG residues involved in the helix-helix interaction. The interchangeability of the LAGLIDADG helix interaction was explored by grafting interfacial residues from the homodimeric I-CreI into the corresponding positions in the monomeric I-DmoI. The resulting LAGLIDADG exchange mutant is partially active, preferring to nick dsDNA rather than making the customary double-strand break. A series of partial revertants within the mutated LAGLIDADG region are shown to restore cleavage activity to varying degrees resulting in one I-DmoI mutant that is more active than wild-type I-DmoI. The phenotype of some of these mutants was reconciled on the basis of similarity to the GxxxG helix interaction found in transmembrane proteins. Additionally, a split variant of I-DmoI was created, demonstrating that the LAGLIDADG helices of I-DmoI are capable of forming and maintaining the protein-protein interface in trans to create an active heterodimer.
Figures




Similar articles
-
Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI.J Mol Biol. 1999 Mar 5;286(4):1123-36. doi: 10.1006/jmbi.1998.2519. J Mol Biol. 1999. PMID: 10047486
-
From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes.J Mol Biol. 2006 Aug 25;361(4):744-54. doi: 10.1016/j.jmb.2006.06.063. Epub 2006 Jul 12. J Mol Biol. 2006. PMID: 16872628
-
Structure and dynamics of mesophilic variants from the homing endonuclease I-DmoI.J Comput Aided Mol Des. 2017 Dec;31(12):1063-1072. doi: 10.1007/s10822-017-0087-5. Epub 2017 Nov 25. J Comput Aided Mol Des. 2017. PMID: 29177929
-
Crystal structure of I-DmoI in complex with its target DNA provides new insights into meganuclease engineering.Proc Natl Acad Sci U S A. 2008 Nov 4;105(44):16888-93. doi: 10.1073/pnas.0804795105. Epub 2008 Oct 30. Proc Natl Acad Sci U S A. 2008. PMID: 18974222 Free PMC article.
-
General classes and functions of four-helix bundle cytokines.Adv Protein Chem. 1998;52:1-65. doi: 10.1016/s0065-3233(08)60432-5. Adv Protein Chem. 1998. PMID: 9917917 Review. No abstract available.
Cited by
-
Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation.Nucleic Acids Res. 2009 Feb;37(3):877-90. doi: 10.1093/nar/gkn1007. Epub 2008 Dec 22. Nucleic Acids Res. 2009. PMID: 19103658 Free PMC article.
-
Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification.Structure. 2011 Jan 12;19(1):7-15. doi: 10.1016/j.str.2010.12.003. Structure. 2011. PMID: 21220111 Free PMC article. Review.
-
Computer design of obligate heterodimer meganucleases allows efficient cutting of custom DNA sequences.Nucleic Acids Res. 2008 Apr;36(7):2163-73. doi: 10.1093/nar/gkn059. Epub 2008 Feb 14. Nucleic Acids Res. 2008. PMID: 18276641 Free PMC article.
-
Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity.Nucleic Acids Res. 2011 Jan;39(1):1-18. doi: 10.1093/nar/gkq742. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805246 Free PMC article. Review.
-
A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences.Nucleic Acids Res. 2006;34(22):e149. doi: 10.1093/nar/gkl720. Epub 2006 Nov 27. Nucleic Acids Res. 2006. PMID: 17130168 Free PMC article.
References
-
- Dalgaard J.Z., Klar,A., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the H-N-H family. Nucleic Acids Res., 25, 4626–4638. - PMC - PubMed
-
- Hensgens L.A.M., Bonen,L., de Haan,M., van der Horst,G. and Grivell,L.A. (1983) Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell, 32, 379–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

