Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells
- PMID: 15194498
- PMCID: PMC7111015
- DOI: 10.1016/j.bbrc.2004.05.107
Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells
Abstract
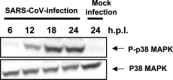
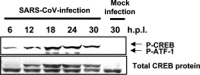
Severe acute respiratory syndrome (SARS) has become a global public health emergency. Understanding the molecular mechanisms of SARS-induced cytopathic effects (CPEs) is a rational approach for the prevention of SARS, and an understanding of the cellular stress responses induced by viral infection is important for understanding the CPEs. Polyclonal antibodies, which recognized nucleocapsid (N) and membrane (M) proteins, detected viral N and M proteins in virus-infected Vero E6 cells at least 6 and 12 h post-infection (h.p.i.), respectively. Furthermore, detection of DNA ladder and cleaved caspase-3 in the virus-infected cells at 24h.p.i. indicated that SARS-CoV infection induced apoptotic cell death. Phosphorylation of p38 MAPK was significantly up-regulated at 18 h.p.i. in SARS-CoV-infected cells. The downstream targets of p38 MAPK, MAPKAPK-2, HSP-27, CREB, and eIF4E were phosphorylated in virus-infected cells. The p38 MAPK inhibitor, SB203580, inhibited effectively phosphorylation of HSP-27, CREB, and eIF4E in SARS-CoV-infected cells. However, viral protein synthesis was not affected by treatment of SB203580.
Figures






References
-
- Rota P.A, Oberste M.S, Monroe S.S, Nix W.A, Campagnoli R, Icenogle J.P, Penaranda S, Bankamp B, Maher K, Chen M.H, Tong S, Tamin A, Lowe L, Frace M, DeRisi J.L, Chen Q, Wang D, Erdman D.D, Peret T.C, Burns C, Ksiazek T.G, Rollin P.E, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus A.D, Drosten C, Pallansch M.A, Anderson L.J, Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. - PubMed
-
- Marra M.A, Jones S.J, Astell C.R, Holt R.A, Brooks-Wilson A, Butterfield Y.S, Khattra J, Asano J.K, Barber S.A, Chan S.Y, Cloutier A, Coughlin S.M, Freeman D, Girn N, Griffith O.L, Leach S.R, Mayo M, McDonald H, Montgomery S.B, Pandoh P.K, Petrescu A.S, Robertson A.G, Schein J.E, Siddiqui A, Smailus D.E, Stott J.M, Yang G.S, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth T.F, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples G.A, Tyler S, Vogrig R, Ward D, Watson B, Brunham R.C, Krajden M, Petric M, Skowronski D.M, Upton C, Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. - PubMed
-
- Garrington T.P, Johnson G.L. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell. Biol. 1999;11:211–218. - PubMed
-
- Whitmarsh A.J, Davis R.J. A central control for cell growth. Nature. 2000;403:255–256. - PubMed
-
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410(6824):37–40. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

