Metal binding by the D,DX35E motif of human immunodeficiency virus type 1 integrase: selective rescue of Cys substitutions by Mn2+ in vitro
- PMID: 15194746
- PMCID: PMC421655
- DOI: 10.1128/JVI.78.13.6715-6722.2004
Metal binding by the D,DX35E motif of human immunodeficiency virus type 1 integrase: selective rescue of Cys substitutions by Mn2+ in vitro
Abstract
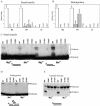
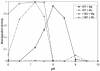
The D,DX(35)E motif characteristic of retroviral integrase enzymes (INs) is expected to bind the required metal cofactors (Mg(2+) or Mn(2+)), but direct evidence for a catalytic role has been lacking. Here we used a metal rescue strategy to investigate metal binding. We established conditions for analysis of an activity of IN, disintegration, in both Mg(2+) and Mn(2+), and tested IN mutants with cysteine substitutions in each acidic residue of the D,DX(35)E motif. Mn(2+) but not Mg(2+) can bind tightly to Cys, so if metal binding at the acidic residues is mechanistically important, it is expected that the Cys-substituted enzymes would be active in the presence of Mn(2+) only. Of the three acidic residues, a strong metal rescue effect was obtained for D116C, a weaker rescue was seen for D64C, and no rescue was seen with E152C. Modest rescue could also be detected for D116C in normal integration in vitro. Comparison to Ser and Ala substitutions at D116 established that the rescue was selective for Cys. Further studies of the response to pH suggest that the metal cofactor may stabilize the deprotonated nucleophile active in catalysis, and studies of the response to NaCl titrations disclose an additional role for the metal cofactor in stabilizing the IN-DNA complex.
Figures


Similar articles
-
Role of metal ions in catalysis by HIV integrase analyzed using a quantitative PCR disintegration assay.Nucleic Acids Res. 2006;34(21):6116-25. doi: 10.1093/nar/gkl862. Epub 2006 Nov 3. Nucleic Acids Res. 2006. PMID: 17085478 Free PMC article.
-
Effects of varying the spacing within the D,D-35-E motif in the catalytic region of retroviral integrase.Virology. 2008 Sep 30;379(2):223-33. doi: 10.1016/j.virol.2008.07.001. Epub 2008 Aug 8. Virology. 2008. PMID: 18687451
-
Divalent cations stimulate preferential recognition of a viral DNA end by HIV-1 integrase.Biochemistry. 1999 Jun 29;38(26):8458-68. doi: 10.1021/bi982870n. Biochemistry. 1999. PMID: 10387092
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
HIV-1 integrase: structural organization, conformational changes, and catalysis.Adv Virus Res. 1999;52:351-69. doi: 10.1016/s0065-3527(08)60306-1. Adv Virus Res. 1999. PMID: 10384242 Review.
Cited by
-
Exploring the Potential of HIV Integrase Inhibitors as Therapeutic Agents Against HSV and HCMV: A Molecular Docking Study.J Exp Pharmacol. 2025 Jul 23;17:507-518. doi: 10.2147/JEP.S524226. eCollection 2025. J Exp Pharmacol. 2025. PMID: 40727394 Free PMC article.
-
Identification of catalytic metal ion ligands in ribozymes.Methods. 2009 Oct;49(2):148-66. doi: 10.1016/j.ymeth.2009.07.005. Epub 2009 Aug 3. Methods. 2009. PMID: 19651216 Free PMC article. Review.
-
Division of labor within human immunodeficiency virus integrase complexes: determinants of catalysis and target DNA capture.J Virol. 2005 Dec;79(24):15376-87. doi: 10.1128/JVI.79.24.15376-15387.2005. J Virol. 2005. PMID: 16306609 Free PMC article.
-
Metal cofactors in the structure and activity of the fowlpox resolvase.J Mol Biol. 2010 May 28;399(1):182-95. doi: 10.1016/j.jmb.2010.03.054. Epub 2010 Apr 7. J Mol Biol. 2010. PMID: 20380839 Free PMC article.
-
Role of metal ions in catalysis by HIV integrase analyzed using a quantitative PCR disintegration assay.Nucleic Acids Res. 2006;34(21):6116-25. doi: 10.1093/nar/gkl862. Epub 2006 Nov 3. Nucleic Acids Res. 2006. PMID: 17085478 Free PMC article.
References
-
- Appa, R. S., C. G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrases in selecting target DNA sites for integration. J. Biol. Chem. 276:45848-45855. - PubMed
-
- Asante-Appiah, E., and A. M. Skalka. 1997. A metal-induced conformational change and activation of HIV-1 integrase. J. Biol. Chem. 272:16196-16205. - PubMed
-
- Bujacz, G., J. Alexandratos, A. Wlodawer, G. Merkel, M. Andrake, R. A. Katz, and A. M. Skalka. 1997. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 272:18161-18168. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

