Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression
- PMID: 15194760
- PMCID: PMC421638
- DOI: 10.1128/JVI.78.13.6846-6854.2004
Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression
Abstract
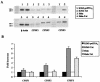
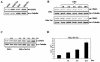
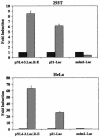
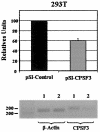
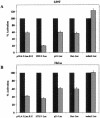
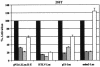
The human immunodeficiency virus type 1 (HIV-1) Tat protein, which is essential for HIV gene expression and viral replication, is known to mediate pleiotropic effects on various cell functions. For instance, Tat protein is able to regulate the rate of transcription of host cellular genes and to interact with the signaling machinery, leading to cellular dysfunction. To study the effect that HIV-1 Tat exerts on the host cell, we identified several genes that were up- or down-regulated in tat-expressing cell lines by using the differential display method. HIV-1 Tat specifically increases the expression of the cleavage and polyadenylation specificity factor (CPSF) 73-kDa subunit (CPSF3) without affecting the expression of the 160- and 100-kDa subunits of the CPSF complex. This complex comprises four subunits and has a key function in the 3'-end processing of pre-mRNAs by a coordinated interaction with other factors. CPSF3 overexpression experiments and knockdown of the endogenous CPSF3 by mRNA interference have shown that this subunit of the complex is an important regulatory protein for both viral and cellular gene expression. In addition to the known CPSF3 function in RNA polyadenylation, we also present evidence that this protein exerts transcriptional activities by repressing the mdm2 gene promoter. Thus, HIV-1-Tat up-regulation of CPSF3 could represent a novel mechanism by which this virus increases mRNA processing, causing an increase in both cell and viral gene expression.
Figures


Similar articles
-
The 73 kDa subunit of the CPSF complex binds to the HIV-1 LTR promoter and functions as a negative regulatory factor that is inhibited by the HIV-1 Tat protein.J Mol Biol. 2007 Sep 14;372(2):317-30. doi: 10.1016/j.jmb.2007.06.075. Epub 2007 Jul 3. J Mol Biol. 2007. PMID: 17669424
-
Modulating HIV-1 replication by RNA interference directed against human transcription elongation factor SPT5.Retrovirology. 2004 Dec 27;1:46. doi: 10.1186/1742-4690-1-46. Retrovirology. 2004. PMID: 15620346 Free PMC article.
-
Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription.Retrovirology. 2006 Aug 7;3:48. doi: 10.1186/1742-4690-3-48. Retrovirology. 2006. PMID: 16893449 Free PMC article.
-
Regulation of expression of human immunodeficiency virus.New Biol. 1990 Jan;2(1):20-31. New Biol. 1990. PMID: 2078551 Review.
-
RNA recognition and regulation of HIV-1 gene expression by viral factor Tat.Biochemistry (Mosc). 1998 May;63(5):489-503. Biochemistry (Mosc). 1998. PMID: 9632883 Review.
Cited by
-
Localization, traffic and function of Rab34 in adipocyte lipid and endocrine functions.J Biomed Sci. 2024 Jan 5;31(1):2. doi: 10.1186/s12929-023-00990-8. J Biomed Sci. 2024. PMID: 38183057 Free PMC article.
-
The retrovirus RNA trafficking granule: from birth to maturity.Retrovirology. 2006 Mar 17;3:18. doi: 10.1186/1742-4690-3-18. Retrovirology. 2006. PMID: 16545126 Free PMC article. Review.
-
Targeting cleavage and polyadenylation specific factor 1 via shRNA inhibits cell proliferation in human ovarian cancer.J Biosci. 2017 Sep;42(3):417-425. doi: 10.1007/s12038-017-9701-x. J Biosci. 2017. PMID: 29358555
-
Multiple Inhibitory Factors Act in the Late Phase of HIV-1 Replication: a Systematic Review of the Literature.Microbiol Mol Biol Rev. 2018 Jan 10;82(1):e00051-17. doi: 10.1128/MMBR.00051-17. Print 2018 Mar. Microbiol Mol Biol Rev. 2018. PMID: 29321222 Free PMC article.
-
Targeting the mRNA endonuclease CPSF73 inhibits breast cancer cell migration, invasion, and self-renewal.iScience. 2022 Jul 20;25(8):104804. doi: 10.1016/j.isci.2022.104804. eCollection 2022 Aug 19. iScience. 2022. PMID: 35992060 Free PMC article.
References
-
- Barak, Y., E. Gottlieb, T. Juven-Gershon, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 8:1739-1749. - PubMed
-
- Benkirane, M., R. F. Chun, H. Xiao, V. V. Ogryzko, B. H. Howard, Y. Nakatani, and K. T. Jeang. 1998. Activation of integrated provirus requires histone acetyltransferase: p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898-24905. - PubMed
-
- Caldwell, R. L., B. S. Egan, and V. L. Shepherd. 2000. HIV-1 Tat represses transcription from the mannose receptor promoter. J. Immunol. 165:7035-7041. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

