Vaccinia virus G1 protein, a predicted metalloprotease, is essential for morphogenesis of infectious virions but not for cleavage of major core proteins
- PMID: 15194761
- PMCID: PMC421688
- DOI: 10.1128/JVI.78.13.6855-6863.2004
Vaccinia virus G1 protein, a predicted metalloprotease, is essential for morphogenesis of infectious virions but not for cleavage of major core proteins
Abstract
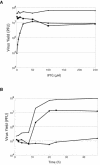
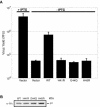
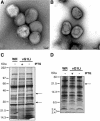
Genes encoding orthologs of the vaccinia virus G1 protein are present in all poxviruses for which sequence information is available, yet neither the role of the protein nor its requirement for virus replication is known. G1 was predicted to be involved in the cleavage of core proteins, based on a transfection study and the presence of an HXXEH motif found in a subset of metallopeptidases. In the present study, we engineered a recombinant vaccinia virus containing a single copy of the G1L gene with a C-terminal epitope tag that is stringently regulated by the Escherichia coli lac repressor. In the absence of inducer, expression of G1 was repressed and virus replication was inhibited. Rescue of infectious virus was achieved by expression of wild-type G1 in trans, but not when the putative protease active site residues histidine-41, glutamate-44, or histidine-45 were mutated. Nevertheless, the synthesis and proteolytic processing of major core and membrane proteins appeared unaffected under nonpermissive conditions, distinguishing the phenotype of the G1L mutant from one in which the gene encoding the I7 protease was repressed. Noninfectious virus particles, assembled in the absence of inducer, did not attain the oval shape or characteristic core structure of mature virions. The polypeptide composition of these particles, however, closely resembled that of wild-type virus. Full-length and shorter forms of the G1 protein were found in the core fraction of virus particles assembled in the presence of inducer, suggesting that G1 is processed by self-cleavage or by another protease.
Figures

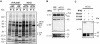


Similar articles
-
Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components.J Virol. 2004 Jun;78(12):6335-43. doi: 10.1128/JVI.78.12.6335-6343.2004. J Virol. 2004. PMID: 15163727 Free PMC article.
-
Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature.J Virol. 2004 Oct;78(19):10238-48. doi: 10.1128/JVI.78.19.10238-10248.2004. J Virol. 2004. PMID: 15367589 Free PMC article.
-
The vaccinia virus G1L putative metalloproteinase is essential for viral replication in vivo.J Virol. 2004 Sep;78(18):9947-53. doi: 10.1128/JVI.78.18.9947-9953.2004. J Virol. 2004. PMID: 15331728 Free PMC article.
-
Immunolocalization of vaccinia virus structural proteins during virion formation.Virology. 1994 Feb;198(2):624-35. doi: 10.1006/viro.1994.1074. Virology. 1994. PMID: 8291244
-
Vaccinia virus proteolysis--a review.Rev Med Virol. 2006 May-Jun;16(3):187-202. doi: 10.1002/rmv.499. Rev Med Virol. 2006. PMID: 16710840 Free PMC article. Review.
Cited by
-
Characterization of a large, proteolytically processed cowpox virus membrane glycoprotein conserved in most chordopoxviruses.Virology. 2015 Sep;483:209-17. doi: 10.1016/j.virol.2015.04.014. Epub 2015 May 15. Virology. 2015. PMID: 25980741 Free PMC article.
-
Interactions of the vaccinia virus A19 protein.J Virol. 2013 Oct;87(19):10710-20. doi: 10.1128/JVI.01261-13. Epub 2013 Jul 24. J Virol. 2013. PMID: 23885084 Free PMC article.
-
Vaccinia virus A21 virion membrane protein is required for cell entry and fusion.J Virol. 2005 Aug;79(15):9458-69. doi: 10.1128/JVI.79.15.9458-9469.2005. J Virol. 2005. PMID: 16014909 Free PMC article.
-
Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis.J Virol. 2009 Sep;83(18):9140-50. doi: 10.1128/JVI.00875-09. Epub 2009 Jul 1. J Virol. 2009. PMID: 19570860 Free PMC article.
-
Vaccinia virus A19 protein participates in the transformation of spherical immature particles to barrel-shaped infectious virions.J Virol. 2013 Oct;87(19):10700-9. doi: 10.1128/JVI.01258-13. Epub 2013 Jul 24. J Virol. 2013. PMID: 23885081 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

