The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness
- PMID: 15194769
- PMCID: PMC421687
- DOI: 10.1128/JVI.78.13.6927-6937.2004
The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness
Abstract
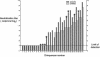
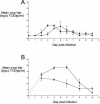
The growth properties and antigenic relatedness of the CAN98-75 (CAN75) and the CAN97-83 (CAN83) human metapneumovirus (HMPV) strains, which represent the two distinct HMPV genetic lineages and exhibit 5 and 63% amino acid divergence in the fusion (F) and attachment (G) proteins, respectively, were investigated in vitro and in rodents and nonhuman primates. Both strains replicated to high titers (> or =6.0 log(10)) in the upper respiratory tract of hamsters and to moderate titers (> or =3.6 log(10)) in the lower respiratory tract. The two lineages exhibited 48% antigenic relatedness based on reciprocal cross-neutralization assay with postinfection hamster sera, and infection with each strain provided a high level of resistance to reinfection with the homologous or heterologous strain. Hamsters immunized with a recombinant human parainfluenza virus type 1 expressing the fusion F protein of the CAN83 strain developed a serum antibody response that efficiently neutralized virus from both lineages and were protected from challenge with either HMPV strain. This result indicates that the HMPV F protein is a major antigenic determinant that mediates extensive cross-lineage neutralization and protection. Both HMPV strains replicated to low titers in the upper and lower respiratory tracts of rhesus macaques but induced high levels of HMPV-neutralizing antibodies in serum effective against both lineages. The level of HMPV replication in chimpanzees was moderately higher, and infected animals developed mild colds. HMPV replicated the most efficiently in the respiratory tracts of African green monkeys, and the infected animals developed a high level of HMPV serum-neutralizing antibodies (1:500 to 1:1,000) effective against both lineages. Reciprocal cross-neutralization assays in which postinfection sera from all three primate species were used indicated that CAN75 and CAN83 are 64 to 99% related antigenically. HMPV-infected chimpanzees and African green monkeys were highly protected from challenge with the heterologous HMPV strain. Taken together, the results from hamsters and nonhuman primates support the conclusion that the two HMPV genetic lineages are highly related antigenically and are not distinct antigenic subtypes or subgroups as defined by reciprocal cross-neutralization in vitro.
Figures
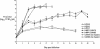

Similar articles
-
An improved plaque reduction virus neutralization assay for human metapneumovirus.J Virol Methods. 2007 Aug;143(2):169-74. doi: 10.1016/j.jviromet.2007.03.005. Epub 2007 Apr 8. J Virol Methods. 2007. PMID: 17420056
-
Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate.J Virol. 2004 Dec;78(23):12877-87. doi: 10.1128/JVI.78.23.12877-12887.2004. J Virol. 2004. PMID: 15542640 Free PMC article.
-
Deletion of M2 gene open reading frames 1 and 2 of human metapneumovirus: effects on RNA synthesis, attenuation, and immunogenicity.J Virol. 2005 Jun;79(11):6588-97. doi: 10.1128/JVI.79.11.6588-6597.2005. J Virol. 2005. PMID: 15890897 Free PMC article.
-
Vaccination approaches to combat human metapneumovirus lower respiratory tract infections.J Clin Virol. 2008 Jan;41(1):49-52. doi: 10.1016/j.jcv.2007.10.022. Epub 2007 Dec 4. J Clin Virol. 2008. PMID: 18054841 Review.
-
Ten years of human metapneumovirus research.J Clin Virol. 2012 Feb;53(2):97-105. doi: 10.1016/j.jcv.2011.10.002. Epub 2011 Nov 9. J Clin Virol. 2012. PMID: 22074934 Review.
Cited by
-
Newly discovered respiratory viruses: significance and implications.Curr Opin Pharmacol. 2007 Oct;7(5):478-83. doi: 10.1016/j.coph.2007.07.004. Epub 2007 Aug 6. Curr Opin Pharmacol. 2007. PMID: 17689145 Free PMC article. Review.
-
Human metapneumovirus persists in BALB/c mice despite the presence of neutralizing antibodies.J Virol. 2004 Dec;78(24):14003-11. doi: 10.1128/JVI.78.24.14003-14011.2004. J Virol. 2004. PMID: 15564507 Free PMC article.
-
A host-range restricted parainfluenza virus type 3 (PIV3) expressing the human metapneumovirus (hMPV) fusion protein elicits protective immunity in African green monkeys.Vaccine. 2005 Feb 25;23(14):1657-67. doi: 10.1016/j.vaccine.2004.10.009. Vaccine. 2005. PMID: 15705469 Free PMC article.
-
Genetic variability of human metapneumovirus amongst an all ages population in Cambodia between 2007 and 2009.Infect Genet Evol. 2013 Apr;15:43-52. doi: 10.1016/j.meegid.2011.01.016. Epub 2011 Feb 1. Infect Genet Evol. 2013. PMID: 21292032 Free PMC article.
-
Novel insights into the host cell glycan binding profile of human metapneumovirus.J Virol. 2024 Jun 13;98(6):e0164123. doi: 10.1128/jvi.01641-23. Epub 2024 May 1. J Virol. 2024. PMID: 38690874 Free PMC article.
References
-
- Bando, H., K. Kondo, M. Kawano, H. Komada, M. Tsurudome, M. Nishio, and Y. Ito. 1990. Molecular cloning and sequence analysis of human parainfluenza type 4A virus HN gene: its irregularities on structure and activities. Virology 175:307-312. - PubMed
-
- Belshe, R. B., L. S. Richardson, W. T. London, D. L. Sly, J. H. Lorfeld, E. Camargo, D. A. Prevar, and R. M. Chanock. 1977. Experimental respiratory syncytial virus infection of four species of primates. J. Med. Virol. 1:157-162. - PubMed
-
- Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

