Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies
- PMID: 15194770
- PMCID: PMC421668
- DOI: 10.1128/JVI.78.13.6938-6945.2004
Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies
Abstract
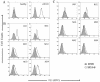
Severe acute respiratory syndrome (SARS) is a life-threatening disease caused by a newly identified coronavirus (CoV), SARS-CoV. The spike (S) glycoprotein of CoV is the major structural protein responsible for induction of host immune response and virus neutralization by antibodies. Hence, knowledge of neutralization determinants on the S protein is helpful for designing protective vaccines. To analyze the antigenic structure of the SARS-CoV S2 domain, the carboxyl-terminal half of the S protein, we first used sera from convalescent SARS patients to test the antigenicity of 12 overlapping fragments spanning the entire S2 and identified two antigenic determinants (Leu 803 to Ala 828 and Pro 1061 to Ser 1093). To determine whether neutralizing antibodies can be elicited by these two determinants, we immunized animals and found that both of them could induce the S2-specific antisera. In some animals, however, only one determinant (Leu 803 to Ala 828) was able to induce the antisera with the binding ability to the native S protein and the neutralizing activity to the SARS-CoV pseudovirus. This determinant is highly conserved across different SARS-CoV isolates. Identification of a conserved antigenic determinant on the S2 domain of the SARS-CoV S protein, which has the potential for inducing neutralizing antibodies, has implications in the development of effective vaccines against SARS-CoV.
Figures


Similar articles
-
Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines.Virology. 2005 Mar 30;334(1):74-82. doi: 10.1016/j.virol.2005.01.034. Virology. 2005. PMID: 15749124 Free PMC article.
-
Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies.J Immunol. 2005 Apr 15;174(8):4908-15. doi: 10.4049/jimmunol.174.8.4908. J Immunol. 2005. PMID: 15814718
-
An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies.J Virol. 2004 Jul;78(13):7217-26. doi: 10.1128/JVI.78.13.7217-7226.2004. J Virol. 2004. PMID: 15194798 Free PMC article.
-
Vaccine design for severe acute respiratory syndrome coronavirus.Viral Immunol. 2005;18(2):327-32. doi: 10.1089/vim.2005.18.327. Viral Immunol. 2005. PMID: 16035944 Review.
-
SARS vaccine development.Emerg Infect Dis. 2005 Jul;11(7):1016-20. doi: 10.3201/1107.050219. Emerg Infect Dis. 2005. PMID: 16022774 Free PMC article. Review.
Cited by
-
Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection.J Infect Dis. 2004 Sep 15;190(6):1119-26. doi: 10.1086/423286. Epub 2004 Aug 2. J Infect Dis. 2004. PMID: 15319862 Free PMC article.
-
Intranasal vaccination of hamsters with a Newcastle disease virus vector expressing the S1 subunit protects animals against SARS-CoV-2 disease.Sci Rep. 2022 Jun 20;12(1):10359. doi: 10.1038/s41598-022-13560-z. Sci Rep. 2022. PMID: 35725862 Free PMC article.
-
Design of a multi-epitope vaccine against SARS-CoV-2 using immunoinformatics approach.Int J Biol Macromol. 2020 Dec 1;164:871-883. doi: 10.1016/j.ijbiomac.2020.07.117. Epub 2020 Jul 15. Int J Biol Macromol. 2020. PMID: 32682041 Free PMC article.
-
A combined nucleocapsid vaccine induces vigorous SARS-CD8+ T-cell immune responses.Genet Vaccines Ther. 2005 Aug 22;3:7. doi: 10.1186/1479-0556-3-7. Genet Vaccines Ther. 2005. PMID: 16115319 Free PMC article.
-
A study on antigenicity and receptor-binding ability of fragment 450-650 of the spike protein of SARS coronavirus.Virology. 2007 Mar 15;359(2):362-70. doi: 10.1016/j.virol.2006.09.022. Epub 2006 Oct 20. Virology. 2007. PMID: 17055551 Free PMC article.
References
-
- Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. - PubMed
-
- Boireau, P., C. Cruciere, and J. Laporte. 1990. Nucleotide sequence of the glycoprotein S gene of bovine enteric coronavirus and comparison with the S proteins of two mouse hepatitis virus strains. J. Gen. Virol. 71:487-492. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

