Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection
- PMID: 15194783
- PMCID: PMC421658
- DOI: 10.1128/JVI.78.13.7069-7078.2004
Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection
Abstract
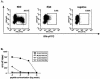
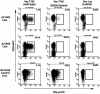
Numerous studies now support that human immunodeficiency virus type 1 (HIV-1) evolution is influenced by immune selection pressure, with population studies showing an association between specific HLA alleles and mutations within defined cytotoxic T-lymphocyte epitopes. Here we combine sequence data and functional studies of CD8 T-cell responses to demonstrate that allele-specific immune pressures also select for mutations flanking CD8 epitopes that impair antigen processing. In persons expressing HLA-A3, we demonstrate consistent selection for a mutation in a C-terminal flanking residue of the normally immunodominant Gag KK9 epitope that prevents its processing and presentation, resulting in a rapid decline in the CD8 T-cell response. This single amino acid substitution also lies within a second HLA-A3-restricted epitope, with the mutation directly impairing recognition by CD8 T cells. Transmission of the mutation to subjects expressing HLA-A3 was shown to prevent the induction of normally immunodominant acute-phase responses to both epitopes. However, subsequent in vivo reversion of the mutation was coincident with delayed induction of new CD8 T-cell responses to both epitopes. These data demonstrate that mutations within the flanking region of an HIV-1 epitope can impair recognition by an established CD8 T-cell response and that transmission of these mutations alters the acute-phase CD8(+) T-cell response. Moreover, reversion of these mutations in the absence of the original immune pressure reveals the potential plasticity of immunologically selected evolutionary changes.
Figures

References
-
- Abele, R., and R. Tampe. 1999. Function of the transport complex TAP in cellular immune recognition. Biochim. Biophys. Acta 1461:405-419. - PubMed
-
- Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyo. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107-112. - PubMed
-
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
-
- Altfeld, M., M. M. Addo, R. L. Eldridge, X. G. Yu, S. Thomas, A. Khatri, D. Strick, M. N. Phillips, G. B. Cohen, S. A. Islam, S. A. Kalams, C. Brander, P. J. Goulder, E. S. Rosenberg, and B. D. Walker. 2001. Vpr is preferentially targeted by CTL during HIV-1 infection. J. Immunol. 167:2743-2752. - PubMed
-
- Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330-7340. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

