Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo
- PMID: 15194785
- PMCID: PMC421671
- DOI: 10.1128/JVI.78.13.7089-7096.2004
Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo
Abstract
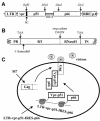
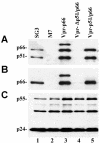
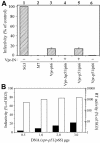
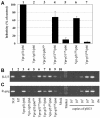
The human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is a heterodimer comprised of two structurally distinct subunits (p51 and p66). Since p51 and p66 are derived from the same coding region, subunit-specific structure-function studies of RT have been conducted exclusively by in vitro biochemical approaches. To study RT subunit function in the context of infectious virus, we constructed an LTR-vpr-p51-IRES-p66 expression cassette in which the HIV-1 vpr gene was fused in frame with p51, followed by an internal ribosome entry site (IRES) sequence and the p66 coding region. By coexpression with RT-deficient proviral DNA, we demonstrated that the p66 subunit is specifically and selectively packaged into virions as a Vpr-p51/p66 complex. Our analysis showed that cleavage by the viral protease liberates Vpr and generates functional heterodimeric RT (p51/p66) that supports HIV-1 reverse transcription and virus infection. By exploiting this novel trans-complementation approach, we demonstrated, for the first time with infectious virions, that the YMDD aspartates of p66 are both required and sufficient for RT polymerase function. Mutational analyses of the p51 YMDD aspartates indicated that they play an important structural role in p51 folding and subunit interactions that are required for the formation of an active RT heterodimer within infected cells. Understanding the role of the individual RT subunits in RNA- and DNA-dependent DNA synthesis is integral to our understanding of RT function. Our findings will lead to important new insights into the role of the p51 and p66 subunits in HIV-1 reverse transcription.
Figures


Similar articles
-
Analysis of human immunodeficiency virus type 1 reverse transcriptase subunit structure/function in the context of infectious virions and human target cells.Antimicrob Agents Chemother. 2005 Sep;49(9):3762-9. doi: 10.1128/AAC.49.9.3762-3769.2005. Antimicrob Agents Chemother. 2005. PMID: 16127051 Free PMC article.
-
Identification of amino acid residues in the human immunodeficiency virus type-1 reverse transcriptase tryptophan-repeat motif that are required for subunit interaction using infectious virions.J Mol Biol. 2005 Jun 17;349(4):673-84. doi: 10.1016/j.jmb.2005.03.057. Epub 2005 Apr 7. J Mol Biol. 2005. PMID: 15893326
-
Analysis of amino acids in the beta7-beta8 loop of human immunodeficiency virus type 1 reverse transcriptase for their role in virus replication.J Mol Biol. 2007 Feb 2;365(5):1368-78. doi: 10.1016/j.jmb.2006.10.089. Epub 2006 Nov 10. J Mol Biol. 2007. PMID: 17141805
-
Human Immunodeficiency Virus type 1 reverse transcriptase.Biol Chem Hoppe Seyler. 1996 Feb;377(2):97-120. Biol Chem Hoppe Seyler. 1996. PMID: 8868066 Review.
-
Buried surface analysis of HIV-1 reverse transcriptase p66/p51 heterodimer and its interaction with dsDNA template/primer.J Mol Recognit. 1994 Jun;7(2):157-61. doi: 10.1002/jmr.300070212. J Mol Recognit. 1994. PMID: 7530020 Review.
Cited by
-
Structural Maturation of HIV-1 Reverse Transcriptase-A Metamorphic Solution to Genomic Instability.Viruses. 2016 Sep 27;8(10):260. doi: 10.3390/v8100260. Viruses. 2016. PMID: 27690082 Free PMC article. Review.
-
Inducible cell lines producing replication-defective human immunodeficiency virus particles containing envelope glycoproteins stabilized in a pretriggered conformation.J Virol. 2024 Dec 17;98(12):e0172024. doi: 10.1128/jvi.01720-24. Epub 2024 Nov 7. J Virol. 2024. PMID: 39508605 Free PMC article.
-
Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay.Mol Cell Biol. 2006 May;26(10):3695-706. doi: 10.1128/MCB.26.10.3695-3706.2006. Mol Cell Biol. 2006. PMID: 16648466 Free PMC article.
-
Inhibition of reverse transcriptase activity increases stability of the HIV-1 core.J Virol. 2013 Jan;87(1):683-7. doi: 10.1128/JVI.01228-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077298 Free PMC article.
-
Murine leukemia virus reverse transcriptase: structural comparison with HIV-1 reverse transcriptase.Virus Res. 2008 Jun;134(1-2):186-202. doi: 10.1016/j.virusres.2008.01.001. Epub 2008 Feb 21. Virus Res. 2008. PMID: 18294720 Free PMC article. Review.
References
-
- Arnold, E., A. Jacobo-Molina, R. G. Nanni, R. L. Williams, X. Lu, J. Ding, A. D. Clark, Jr., A. Zhang, A. L. Ferris, P. Clark, et al. 1992. Structure of HIV-1 reverse transcriptase/DNA complex at 7 Å resolution showing active site locations. Nature 357:85-89. - PubMed
-
- Arts, E. J., X. Li, Z. Gu, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNA(Lys-3) as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. - PubMed
-
- Baillon, J. G., N. T. Nashed, A. Kumar, S. H. Wilson, and D. M. Jerina. 1991. A leucine zipper-like motif may mediate HIV reverse transcriptase subunit binding. New Biol. 3:1015-1019. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

