Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir
- PMID: 15194788
- PMCID: PMC421656
- DOI: 10.1128/JVI.78.13.7124-7130.2004
Mutations in the human cytomegalovirus UL27 gene that confer resistance to maribavir
Abstract
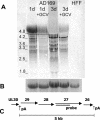
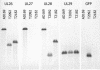
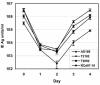
Previous drug selection experiments resulted in the isolation of a human cytomegalovirus (CMV) UL97 phosphotransferase mutant resistant to the benzimidazole compound maribavir (1263W94), reflecting the anti-UL97 effect of this drug. Three other CMV strains were plaque purified during these experiments. These strains showed lower-grade resistance to maribavir than the UL97 mutant. Extensive DNA sequence analyses showed no changes from the baseline strain AD169 in UL97, the genes involved in DNA replication, and most structural proteins. However, changes were identified in UL27 where each strain contained a different mutation (R233S, W362R, or a combination of A406V and a stop at codon 415). The mutation at codon 415 is predicted to truncate the expressed UL27 protein by 193 codons (32% of UL27) with a loss of nuclear localization. The expression of full-length UL27 as a green fluorescent fusion protein in uninfected fibroblasts resulted in nuclear and nucleolar fluorescence, whereas cytoplasmic localization was observed when codons 1 to 415 were similarly expressed. Viable UL27 deletion mutants were created by recombination and showed slight growth attenuation and maribavir resistance in cell culture. Marker transfer experiments confirmed that UL27 mutations conferred maribavir resistance. The UL27 sequence was well conserved in a sample of 16 diverse clinical isolates. Mutation in UL27, a betaherpesvirus-specific early gene of unknown biological function, may adapt the virus for growth in the absence of UL97 activity.
Figures


References
-
- Baek M. C., P. M. Krosky, Z. He, and D. M. Coen. 2002. Specific phosphorylation of exogenous protein and peptide substrates by the human cytomegalovirus UL97 protein kinase. Importance of the P+5 position. J. Biol. Chem. 277:29593-29599. - PubMed
-
- Biron K. K., R. J. Harvey, S. C. Chamberlain, S. S. Good, A. A. Smith III, M. G. Davis, C. L. Talarico, W. H. Miller, R. Ferris, R. E. Dornsife, S. C. Stanat, J. C. Drach, L. B. Townsend, and G. W. Koszalka. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365-2372. - PMC - PubMed
-
- Chou S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

